scRNAseq_combined_05_visualization
retogerber
2024-02-12
Last updated: 2024-02-12
Checks: 7 0
Knit directory: synovialscrnaseq/
This reproducible R Markdown analysis was created with workflowr (version 1.6.2). The Checks tab describes the reproducibility checks that were applied when the results were created. The Past versions tab lists the development history.
Great! Since the R Markdown file has been committed to the Git repository, you know the exact version of the code that produced these results.
Great job! The global environment was empty. Objects defined in the global environment can affect the analysis in your R Markdown file in unknown ways. For reproduciblity it’s best to always run the code in an empty environment.
The command set.seed(20210105) was run prior to running the code in the R Markdown file. Setting a seed ensures that any results that rely on randomness, e.g. subsampling or permutations, are reproducible.
Great job! Recording the operating system, R version, and package versions is critical for reproducibility.
Nice! There were no cached chunks for this analysis, so you can be confident that you successfully produced the results during this run.
Great job! Using relative paths to the files within your workflowr project makes it easier to run your code on other machines.
Great! You are using Git for version control. Tracking code development and connecting the code version to the results is critical for reproducibility.
The results in this page were generated with repository version 58eeb06. See the Past versions tab to see a history of the changes made to the R Markdown and HTML files.
Note that you need to be careful to ensure that all relevant files for the analysis have been committed to Git prior to generating the results (you can use wflow_publish or wflow_git_commit). workflowr only checks the R Markdown file, but you know if there are other scripts or data files that it depends on. Below is the status of the Git repository when the results were generated:
Ignored files:
Ignored: '/
Ignored: .Rhistory
Ignored: .Rproj.user/
Ignored: .empty/
Ignored: analysis/.Rhistory
Ignored: analysis/iSEE_interactive_document.html
Ignored: code/test_files/
Ignored: data/Culemann/
Ignored: data/E-MTAB-8322/
Ignored: data/Synovial scRNA-seq samples - Sheet1.csv
Ignored: data/Zhang_top20_singlecell_cluster_markers_fromGithub.csv
Ignored: data/findMarkers_results.rds
Ignored: data/findMarkers_results_v2.rds
Ignored: data/info/
Ignored: data/syn_sce_tidy_filtered.rds
Ignored: data/syn_sce_tidy_hvg.rds
Ignored: data/syn_sce_tidy_hvg_cms.rds
Ignored: docs/
Ignored: output/Figures_Paper/
Ignored: output/Sample_summaries_RA_comparisons.rds
Ignored: output/Sample_summaries_direct_dissociation.rds
Ignored: output/Sample_summaries_exvivo_treatment.rds
Ignored: output/Suppl_Figure_4d.rds
Ignored: output/barcodes.txt
Ignored: output/barcodes_filtered.txt
Ignored: output/column_metadata_filtered.txt
Ignored: output/combined_v7_SingleR_markers.rds
Ignored: output/combined_v7_SingleR_predictions.rds
Ignored: output/combined_v7_SingleR_predictions_lclc.rds
Ignored: output/combined_v7_SingleR_predictions_reclc.rds
Ignored: output/combined_v7_SingleR_predictions_recrec.rds
Ignored: output/combined_v7_SingleR_trained.rds
Ignored: output/combined_v7_sce.rds
Ignored: output/combined_v7_sce_filtered.rds
Ignored: output/combined_v7_sce_hvg.rds
Ignored: output/combined_v7_sce_hvg_cms.rds
Ignored: output/combined_v7_sce_hvg_cms_annotated.rds
Ignored: output/combined_v7_sce_tmp_hvg_cms.rds
Ignored: output/combined_v7_upsetplot_genelists.rds
Ignored: output/count_matrix_filtered.mtx
Ignored: output/count_matrix_unfiltered.mtx
Ignored: output/emptyDrops_result_v4.rds
Ignored: output/emptyDrops_result_v4_tmp.rds
Ignored: output/emptyDrops_result_v4tmptmp.rds
Ignored: output/findMarkers_results_v6.rds
Ignored: output/findMarkers_results_v6_ec.rds
Ignored: output/findMarkers_results_v6_main.rds
Ignored: output/findMarkers_results_v6_mp.rds
Ignored: output/findMarkers_results_v6_sf.rds
Ignored: output/findMarkers_results_v6_tc.rds
Ignored: output/findMarkers_results_v7_ec.rds
Ignored: output/findMarkers_results_v7_main.rds
Ignored: output/findMarkers_results_v7_mp.rds
Ignored: output/findMarkers_results_v7_sf.rds
Ignored: output/findMarkers_results_v7_tc.rds
Ignored: output/genes.txt
Ignored: output/genes_filtered.txt
Ignored: output/goana_results_v6_ec.rds
Ignored: output/goana_results_v6_mp.rds
Ignored: output/preprocessing_number_of_cells.rds
Ignored: output/syn_v4_sce_emptyDrops_invivo.rds
Ignored: output/syn_v4_swappedDrops_24300_after.rds
Ignored: output/syn_v4_swappedDrops_24300_before.rds
Ignored: output/syn_v4_swappedDrops_24793_after.rds
Ignored: output/syn_v4_swappedDrops_24793_before.rds
Ignored: output/syn_v6_cluster_cellid_match_invivo.rds
Ignored: output/syn_v6_clustering_lookup_invivo.rds
Ignored: output/syn_v6_clustering_lookup_multiple_invivo.rds
Ignored: output/syn_v6_sce.rds
Ignored: output/syn_v6_sce_Figure8.rds
Ignored: output/syn_v6_sce_Figure8_dic_ls.rds
Ignored: output/syn_v6_sce_ec_invivo.rds
Ignored: output/syn_v6_sce_filtered_invivo.rds
Ignored: output/syn_v6_sce_hdf5/
Ignored: output/syn_v6_sce_hvg_cms_doublet_invivo.rds
Ignored: output/syn_v6_sce_hvg_cms_doublet_subcluster_invivo.rds
Ignored: output/syn_v6_sce_hvg_invivo.rds
Ignored: output/syn_v6_sce_hvg_marker_genes.rds
Ignored: output/syn_v6_sce_mp_invivo.rds
Ignored: output/syn_v6_sce_sf_invivo.rds
Ignored: output/syn_v6_sce_tc_invivo.rds
Ignored: output/syn_v6_sfig1.rds
Ignored: output/syn_v6_vst_out_invivo.rds
Ignored: output/syn_v7_cluster_cellid_match_invivo.rds
Ignored: output/syn_v7_clustering_lookup_invivo.rds
Ignored: output/syn_v7_clustering_lookup_multiple_invivo.rds
Ignored: output/syn_v7_sce.rds
Ignored: output/syn_v7_sce_Figure8.rds
Ignored: output/syn_v7_sce_Figure8_dic_ls.rds
Ignored: output/syn_v7_sce_ec_invivo.rds
Ignored: output/syn_v7_sce_ec_invivo_trajectory.rds
Ignored: output/syn_v7_sce_ec_invivo_trajectory2.rds
Ignored: output/syn_v7_sce_ec_invivo_trajectory2_ATres.rds
Ignored: output/syn_v7_sce_ec_invivo_trajectory_icMat.rds
Ignored: output/syn_v7_sce_filtered_invivo.rds
Ignored: output/syn_v7_sce_hdf5/
Ignored: output/syn_v7_sce_hvg_cms_doublet_invivo.rds
Ignored: output/syn_v7_sce_hvg_cms_doublet_subcluster_invivo.rds
Ignored: output/syn_v7_sce_hvg_cms_doublet_subcluster_invivo_cleaned.rds
Ignored: output/syn_v7_sce_hvg_invivo.rds
Ignored: output/syn_v7_sce_mp_invivo.rds
Ignored: output/syn_v7_sce_sf_invivo.rds
Ignored: output/syn_v7_sce_tc_invivo.rds
Ignored: output/syn_v7_sfig1.rds
Ignored: output/syn_v7_vst_out_invivo.rds
Untracked files:
Untracked: analysis/clean_and_save_sce.R
Untracked: analysis/description_integration_wei_stephenson
Untracked: analysis/scRNAseq_complete_01_preprocessing_comparison.Rmd
Untracked: analysis/scRNAseq_complete_05_ec_trajectory_analysis.Rmd
Untracked: analysis/scRNAseq_complete_05_ec_trajectory_analysis_2.Rmd
Untracked: analysis/scRNAseq_complete_05_ec_trajectory_analysis_3.Rmd
Untracked: code/plot_utilities.Rmd
Untracked: code/rebuild_ezRun.R
Untracked: code/tmp1.R
Untracked: code/tmp1.Rmd
Untracked: nonhosted_public/
Untracked: singRstudio.sh.bak
Unstaged changes:
Modified: analysis/scRNAseq_combined_06_Figures.Rmd
Modified: analysis/scRNAseq_complete_04-2_celltype_markers.Rmd
Modified: analysis/scRNAseq_complete_04-2_celltype_markers_subcelltypes.Rmd
Modified: analysis/scRNAseq_complete_04_Annotation_v7.Rmd
Modified: analysis/scRNAseq_complete_Figures.Rmd
Modified: analysis/write_tsv.Rmd
Modified: code/create_hdf5.R
Note that any generated files, e.g. HTML, png, CSS, etc., are not included in this status report because it is ok for generated content to have uncommitted changes.
These are the previous versions of the repository in which changes were made to the R Markdown (analysis/scRNAseq_combined_05_visualization.Rmd) and HTML (public/scRNAseq_combined_05_visualization.html) files. If you’ve configured a remote Git repository (see ?wflow_git_remote), click on the hyperlinks in the table below to view the files as they were in that past version.
| File | Version | Author | Date | Message |
|---|---|---|---|---|
| Rmd | 58eeb06 | Reto Gerber | 2023-05-30 | add new version |
| html | 58eeb06 | Reto Gerber | 2023-05-30 | add new version |
Set up
suppressPackageStartupMessages({
library(magrittr)
library(SingleCellExperiment)
library(BiocParallel)
library(ggplot2)
})
n_workers <- 10
RhpcBLASctl::blas_set_num_threads(n_workers)
bpparam <- BiocParallel::MulticoreParam(workers=n_workers, RNGseed = 123)
analysis_version <- 7
here::here()[1] "/home/retger/Synovial/synovialscrnaseq"source(here::here("code","utilities_plots.R"))
set.seed(100)syn_sce <- readRDS(file = here::here("output",paste0("combined_v",analysis_version,"_sce_hvg_cms.rds")))predictions <- readRDS(here::here("output",paste0("combined_v",analysis_version,"_SingleR_predictions_recrec.rds")))
syn_ref <- syn_sce[,!(syn_sce$Protocol %in% c("wei","stephenson"))]Loading required package: tidySingleCellExperiment
Attaching package: 'tidySingleCellExperiment'The following object is masked from 'package:IRanges':
sliceThe following object is masked from 'package:S4Vectors':
renameThe following object is masked from 'package:matrixStats':
countThe following object is masked from 'package:magrittr':
extractThe following object is masked from 'package:stats':
filterLoading required package: BiocSingularlookup <- unique(colData(syn_ref)[,c("minor_celltype","main_celltype")])
mainlabsdf <- dplyr::left_join(
data.frame(minor_celltype=predictions$labels),
as.data.frame(lookup),
by="minor_celltype")Number/proportion of celltypes in stephenson and wei.
sce_sub <- syn_sce[,syn_sce$Protocol %in% c("stephenson","wei")]
sce_sub$minor_celltype <- predictions$labels#[syn_sce$Protocol %in% c("stephenson","wei")]
sce_sub$main_celltype <- mainlabsdf$main_celltype#[syn_sce$Protocol %in% c("stephenson","wei")]
print("main_celltype")[1] "main_celltype"table(sce_sub$main_celltype, sce_sub$Protocol)
stephenson wei
B cells 470 1
Dendritic cells 209 2
Endothelial cells 623 4055
Fibroblasts 8130 14838
Macrophages 3766 280
Mast cells 134 0
Neutrophils 2 0
Pericytes/Mural cells 45 812
Plasmablasts 251 0
Plasmacytoid DCs 17 2
T cells/NK cells 5406 22tab_ste <- table(sce_sub$main_celltype[sce_sub$Protocol=="stephenson"])
tab_wei <- table(sce_sub$main_celltype[sce_sub$Protocol=="wei"])
print("Stephenson")[1] "Stephenson"round(tab_ste/sum(tab_ste),3)
B cells Dendritic cells Endothelial cells
0.025 0.011 0.033
Fibroblasts Macrophages Mast cells
0.427 0.198 0.007
Neutrophils Pericytes/Mural cells Plasmablasts
0.000 0.002 0.013
Plasmacytoid DCs T cells/NK cells
0.001 0.284 print("Wei")[1] "Wei"round(tab_wei/sum(tab_wei),3)
B cells Dendritic cells Endothelial cells
0.000 0.000 0.203
Fibroblasts Macrophages Pericytes/Mural cells
0.741 0.014 0.041
Plasmacytoid DCs T cells/NK cells
0.000 0.001 print("minor_celltype")[1] "minor_celltype"table(sce_sub$minor_celltype, sce_sub$Protocol)
stephenson
ACKRhigh IL1R1+ CLU+ SELE+ venous 84
ACKRhigh IL1R1+ CLU+ SELEhigh TNFAIP3+ venous 126
ACKRhigh IL1R1+ CLU+ VCAN+ venous 125
ACKRmed CLU- SPARChigh 71
B cells 470
C1QA/B/C+ FOLR2low CCR2+ CD48+ CLEC10A 441
CADM1high ACAN+ DKK3+ 376
CCR7high LEF1+ SELL+ 743
CCR7med LEF1low SELLlow 573
CD1C+ CLEC10A+ 99
CD3- NKG7- KLRB1+ IL7R+ 35
CD3- NKG7+ GNLY+ 268
CD48+ CLEC10A+ 159
CD48high S100A12+ IL1B+ 38
CD48low SPP1+ 235
CLEC9A+ CADM1+ CLNK+ 56
Endothelial cells 49
FOLR2+ MERTK+ TIMD4+ & FOLR2low MERTKlow SPP1+ subsets 630
FOLR2high MERTK+ SELENOPhigh CD48med 571
FOLR2high MERTK+ SELENOPhigh COLEC12high LYVE1+ CD209+ SLC40A1+ 436
FOLR2high MERTK+ SELENOPhigh COLEC12high TIMD4+ 713
FOLR2low MERTKlow TOP2A+ CENPF+ proliferating 72
GGT5high CXCL12 high FGF7+ 788
GJA4+ CLDN5+ arterial 39
GZMB- GZMH- GZMK+ 678
GZMB+ GZMH+ GZMK- GNLY+ 287
GZMB+ GZMH+ GZMK+ 1387
HLA-DRAhigh CD74+ 2320
IDO1+ LAMP3+ 54
KDR+ SPP1+ SPARChigh capillary 89
LYVE1+ PROX1+ CCL21+ lymphatic 30
Macrophages 471
Mast cells 134
MMP13+ 478
Neutrophils 2
Pericytes/Mural cells 45
Plasmablasts 251
Plasmacytoid DCs 17
PRG4+ CD55+ TWISTNB+ lining SF 3083
SERPINE1+ COL5A3+ LOXL2high 349
TIGIT+ CTLA4+ 1305
TNXBhigh IGFBP6+ FGFBP2+ 736
TOP2A+ CENPF+ 130
TOP2A+ CENPF+ proliferating 10
wei
ACKRhigh IL1R1+ CLU+ SELE+ venous 631
ACKRhigh IL1R1+ CLU+ SELEhigh TNFAIP3+ venous 630
ACKRhigh IL1R1+ CLU+ VCAN+ venous 997
ACKRmed CLU- SPARChigh 854
B cells 1
C1QA/B/C+ FOLR2low CCR2+ CD48+ CLEC10A 1
CADM1high ACAN+ DKK3+ 1676
CCR7high LEF1+ SELL+ 0
CCR7med LEF1low SELLlow 1
CD1C+ CLEC10A+ 1
CD3- NKG7- KLRB1+ IL7R+ 0
CD3- NKG7+ GNLY+ 9
CD48+ CLEC10A+ 13
CD48high S100A12+ IL1B+ 13
CD48low SPP1+ 25
CLEC9A+ CADM1+ CLNK+ 1
Endothelial cells 1
FOLR2+ MERTK+ TIMD4+ & FOLR2low MERTKlow SPP1+ subsets 45
FOLR2high MERTK+ SELENOPhigh CD48med 33
FOLR2high MERTK+ SELENOPhigh COLEC12high LYVE1+ CD209+ SLC40A1+ 62
FOLR2high MERTK+ SELENOPhigh COLEC12high TIMD4+ 70
FOLR2low MERTKlow TOP2A+ CENPF+ proliferating 9
GGT5high CXCL12 high FGF7+ 4716
GJA4+ CLDN5+ arterial 291
GZMB- GZMH- GZMK+ 3
GZMB+ GZMH+ GZMK- GNLY+ 0
GZMB+ GZMH+ GZMK+ 2
HLA-DRAhigh CD74+ 1946
IDO1+ LAMP3+ 0
KDR+ SPP1+ SPARChigh capillary 562
LYVE1+ PROX1+ CCL21+ lymphatic 31
Macrophages 9
Mast cells 0
MMP13+ 1676
Neutrophils 0
Pericytes/Mural cells 812
Plasmablasts 0
Plasmacytoid DCs 2
PRG4+ CD55+ TWISTNB+ lining SF 1729
SERPINE1+ COL5A3+ LOXL2high 592
TIGIT+ CTLA4+ 1
TNXBhigh IGFBP6+ FGFBP2+ 2503
TOP2A+ CENPF+ 6
TOP2A+ CENPF+ proliferating 58tab_ste <- table(sce_sub$minor_celltype[sce_sub$Protocol=="stephenson"])
tab_wei <- table(sce_sub$minor_celltype[sce_sub$Protocol=="wei"])
print("Stephenson")[1] "Stephenson"round(tab_ste/sum(tab_ste),3)
ACKRhigh IL1R1+ CLU+ SELE+ venous
0.004
ACKRhigh IL1R1+ CLU+ SELEhigh TNFAIP3+ venous
0.007
ACKRhigh IL1R1+ CLU+ VCAN+ venous
0.007
ACKRmed CLU- SPARChigh
0.004
B cells
0.025
C1QA/B/C+ FOLR2low CCR2+ CD48+ CLEC10A
0.023
CADM1high ACAN+ DKK3+
0.020
CCR7high LEF1+ SELL+
0.039
CCR7med LEF1low SELLlow
0.030
CD1C+ CLEC10A+
0.005
CD3- NKG7- KLRB1+ IL7R+
0.002
CD3- NKG7+ GNLY+
0.014
CD48+ CLEC10A+
0.008
CD48high S100A12+ IL1B+
0.002
CD48low SPP1+
0.012
CLEC9A+ CADM1+ CLNK+
0.003
Endothelial cells
0.003
FOLR2+ MERTK+ TIMD4+ & FOLR2low MERTKlow SPP1+ subsets
0.033
FOLR2high MERTK+ SELENOPhigh CD48med
0.030
FOLR2high MERTK+ SELENOPhigh COLEC12high LYVE1+ CD209+ SLC40A1+
0.023
FOLR2high MERTK+ SELENOPhigh COLEC12high TIMD4+
0.037
FOLR2low MERTKlow TOP2A+ CENPF+ proliferating
0.004
GGT5high CXCL12 high FGF7+
0.041
GJA4+ CLDN5+ arterial
0.002
GZMB- GZMH- GZMK+
0.036
GZMB+ GZMH+ GZMK- GNLY+
0.015
GZMB+ GZMH+ GZMK+
0.073
HLA-DRAhigh CD74+
0.122
IDO1+ LAMP3+
0.003
KDR+ SPP1+ SPARChigh capillary
0.005
LYVE1+ PROX1+ CCL21+ lymphatic
0.002
Macrophages
0.025
Mast cells
0.007
MMP13+
0.025
Neutrophils
0.000
Pericytes/Mural cells
0.002
Plasmablasts
0.013
Plasmacytoid DCs
0.001
PRG4+ CD55+ TWISTNB+ lining SF
0.162
SERPINE1+ COL5A3+ LOXL2high
0.018
TIGIT+ CTLA4+
0.068
TNXBhigh IGFBP6+ FGFBP2+
0.039
TOP2A+ CENPF+
0.007
TOP2A+ CENPF+ proliferating
0.001 print("Wei")[1] "Wei"round(tab_wei/sum(tab_wei),3)
ACKRhigh IL1R1+ CLU+ SELE+ venous
0.032
ACKRhigh IL1R1+ CLU+ SELEhigh TNFAIP3+ venous
0.031
ACKRhigh IL1R1+ CLU+ VCAN+ venous
0.050
ACKRmed CLU- SPARChigh
0.043
B cells
0.000
C1QA/B/C+ FOLR2low CCR2+ CD48+ CLEC10A
0.000
CADM1high ACAN+ DKK3+
0.084
CCR7med LEF1low SELLlow
0.000
CD1C+ CLEC10A+
0.000
CD3- NKG7+ GNLY+
0.000
CD48+ CLEC10A+
0.001
CD48high S100A12+ IL1B+
0.001
CD48low SPP1+
0.001
CLEC9A+ CADM1+ CLNK+
0.000
Endothelial cells
0.000
FOLR2+ MERTK+ TIMD4+ & FOLR2low MERTKlow SPP1+ subsets
0.002
FOLR2high MERTK+ SELENOPhigh CD48med
0.002
FOLR2high MERTK+ SELENOPhigh COLEC12high LYVE1+ CD209+ SLC40A1+
0.003
FOLR2high MERTK+ SELENOPhigh COLEC12high TIMD4+
0.003
FOLR2low MERTKlow TOP2A+ CENPF+ proliferating
0.000
GGT5high CXCL12 high FGF7+
0.236
GJA4+ CLDN5+ arterial
0.015
GZMB- GZMH- GZMK+
0.000
GZMB+ GZMH+ GZMK+
0.000
HLA-DRAhigh CD74+
0.097
KDR+ SPP1+ SPARChigh capillary
0.028
LYVE1+ PROX1+ CCL21+ lymphatic
0.002
Macrophages
0.000
MMP13+
0.084
Pericytes/Mural cells
0.041
Plasmacytoid DCs
0.000
PRG4+ CD55+ TWISTNB+ lining SF
0.086
SERPINE1+ COL5A3+ LOXL2high
0.030
TIGIT+ CTLA4+
0.000
TNXBhigh IGFBP6+ FGFBP2+
0.125
TOP2A+ CENPF+
0.000
TOP2A+ CENPF+ proliferating
0.003 set.seed(123)
shuffle <- sample(seq_len(dim(sce_sub)[2]))
scater::plotReducedDim(sce_sub[,shuffle], "UMAP_corrected", colour_by = "minor_celltype",text_by="minor_celltype", other_fields=list("Protocol")) +
guides(color=guide_legend(ncol=1))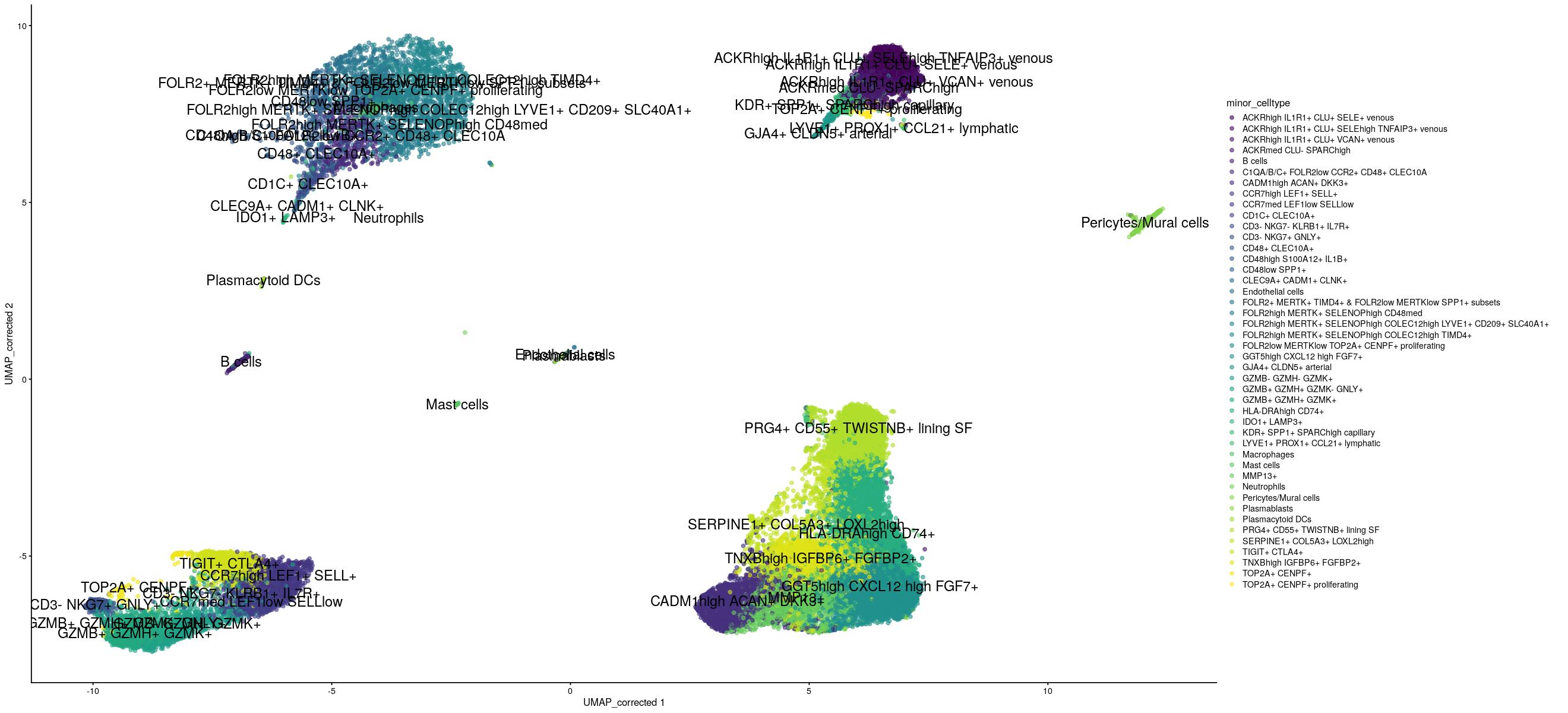
| Version | Author | Date |
|---|---|---|
| 58eeb06 | Reto Gerber | 2023-05-30 |
assign labels to dataset
syn_sce$minor_celltype[syn_sce$Protocol %in% c("wei","stephenson")] <- predictions$labels
syn_sce$main_celltype[syn_sce$Protocol %in% c("wei","stephenson")] <- mainlabsdf$main_celltypesaveRDS(syn_sce,file = here::here("output",paste0("combined_v",analysis_version,"_sce_hvg_cms_annotated.rds")))tab <- table(syn_sce$main_celltype,syn_sce$Protocol)
tab
Protocol_1 Protocol_2 stephenson wei
B cells 367 1186 470 1
Dendritic cells 656 1037 209 2
Endothelial cells 2303 7092 623 4055
Fibroblasts 15596 14836 8130 14838
Macrophages 12932 21744 3766 280
Mast cells 133 171 134 0
Neutrophils 32 394 2 0
Pericytes/Mural cells 161 606 45 812
Plasmablasts 71 102 251 0
Plasmacytoid DCs 40 130 17 2
T cells/NK cells 7358 15811 5406 22as.data.frame(tab) %>%
dplyr::group_by(Var2) %>%
dplyr::mutate(Proportion=Freq/sum(Freq)) %>%
ggplot() +
geom_col(aes(Var2,Proportion,fill=Var1)) +
labs(x=NULL,fill="Cell Type")+
scale_fill_viridis_d()+
main_plot_theme()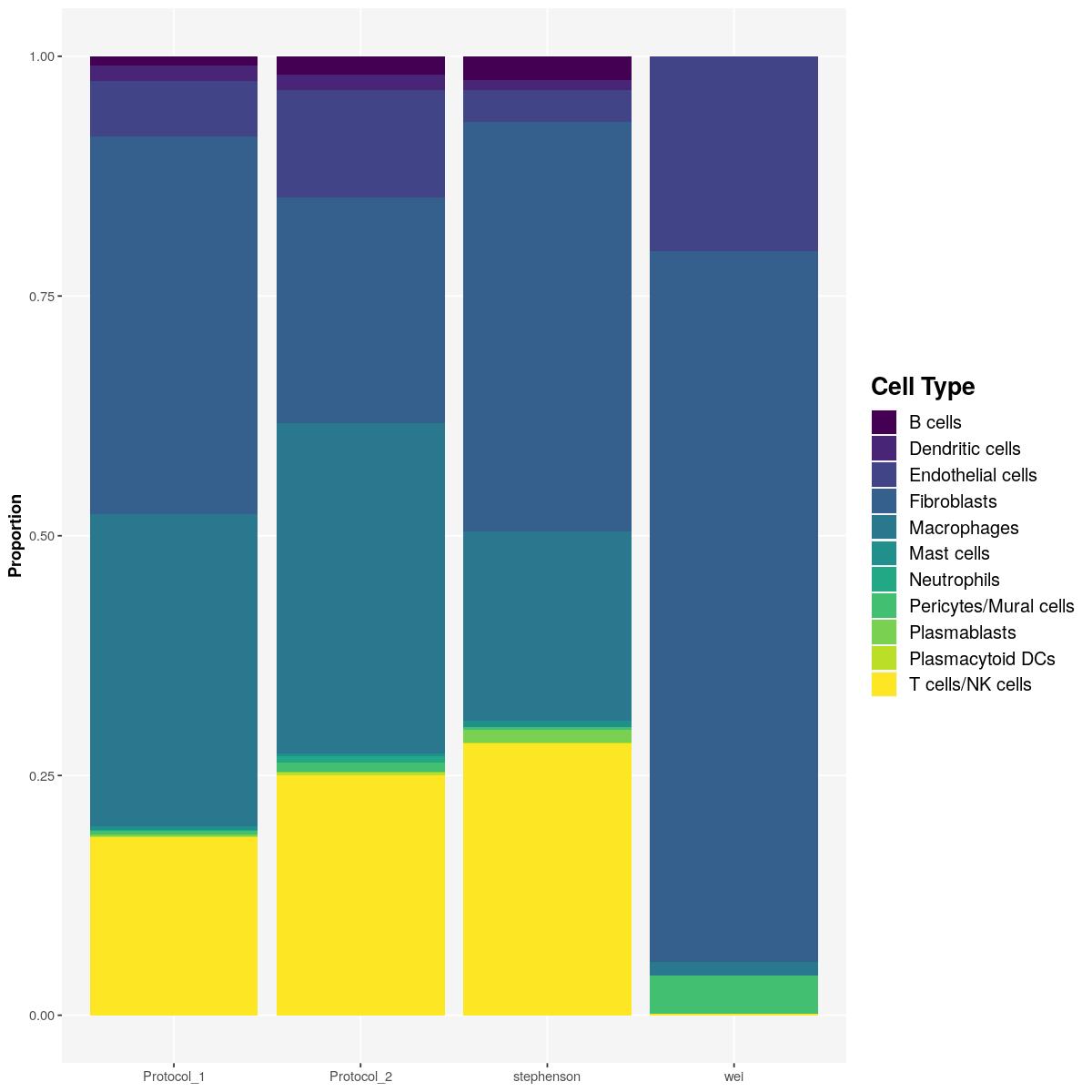
| Version | Author | Date |
|---|---|---|
| 58eeb06 | Reto Gerber | 2023-05-30 |
set.seed(123)
shuffle <- sample(seq_len(dim(syn_sce)[2]))
sce_shuffle <- syn_sce[,shuffle]scater::plotReducedDim(sce_shuffle, "UMAP_corrected", colour_by = "main_celltype",other_fields=list("Protocol"),point_alpha=0.9) +
facet_wrap(~Protocol) +
scale_color_viridis_d()+
main_plot_theme() +
labs(colour="Cell Type",x="UMAP 1", y="UMAP 2")Scale for 'colour' is already present. Adding another scale for 'colour',
which will replace the existing scale.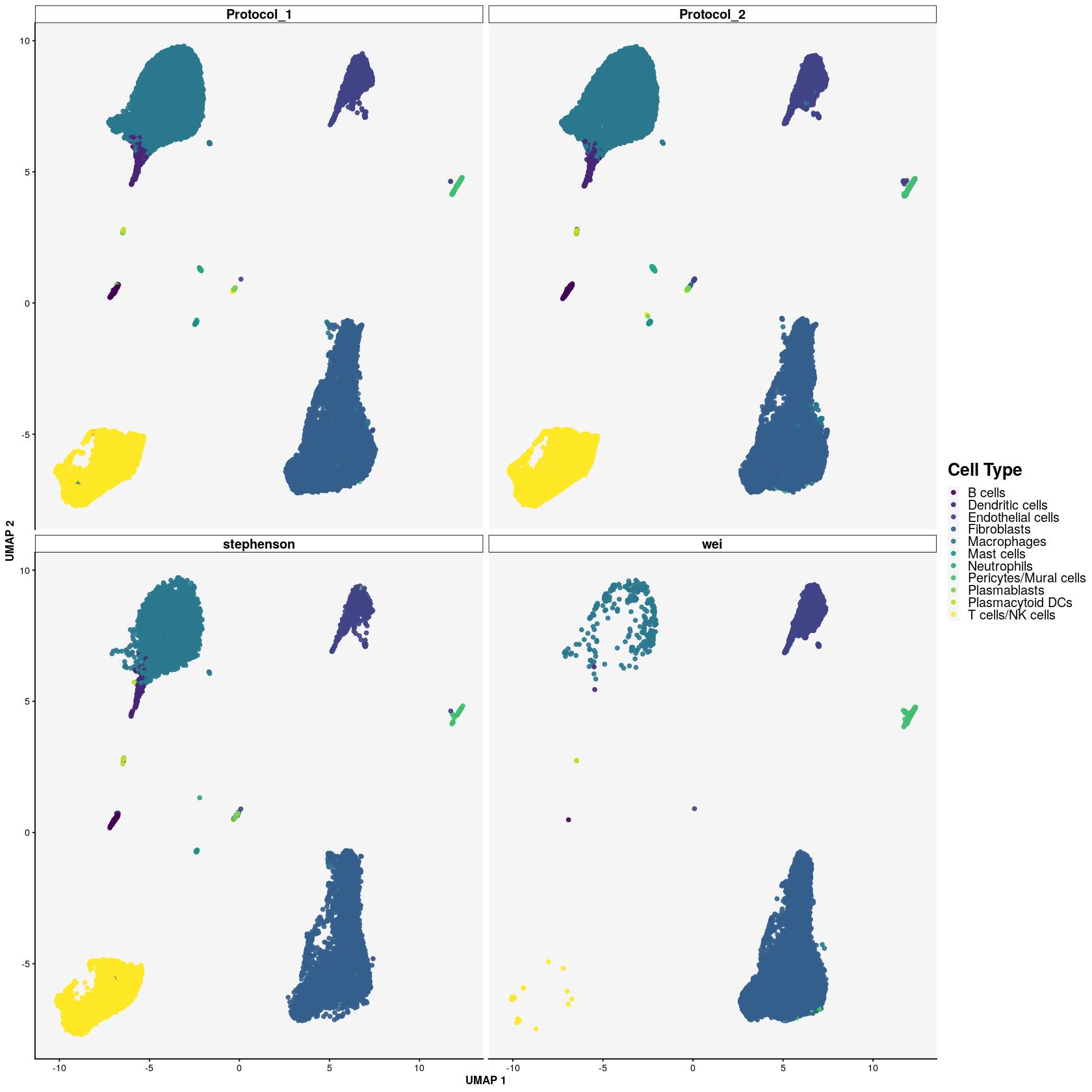
| Version | Author | Date |
|---|---|---|
| 58eeb06 | Reto Gerber | 2023-05-30 |
sessionInfo()R version 4.0.3 (2020-10-10)
Platform: x86_64-pc-linux-gnu (64-bit)
Running under: Ubuntu 20.04 LTS
Matrix products: default
BLAS/LAPACK: /usr/lib/x86_64-linux-gnu/openblas-pthread/libopenblasp-r0.3.8.so
locale:
[1] LC_CTYPE=en_US.UTF-8 LC_NUMERIC=C
[3] LC_TIME=en_US.UTF-8 LC_COLLATE=en_US.UTF-8
[5] LC_MONETARY=en_US.UTF-8 LC_MESSAGES=C
[7] LC_PAPER=en_US.UTF-8 LC_NAME=C
[9] LC_ADDRESS=C LC_TELEPHONE=C
[11] LC_MEASUREMENT=en_US.UTF-8 LC_IDENTIFICATION=C
attached base packages:
[1] parallel stats4 stats graphics grDevices utils datasets
[8] methods base
other attached packages:
[1] gdtools_0.2.3 BiocSingular_1.6.0
[3] tidySingleCellExperiment_1.0.0 ggplot2_3.3.3
[5] BiocParallel_1.24.1 SingleCellExperiment_1.12.0
[7] SummarizedExperiment_1.20.0 Biobase_2.50.0
[9] GenomicRanges_1.42.0 GenomeInfoDb_1.26.7
[11] IRanges_2.24.1 S4Vectors_0.28.1
[13] BiocGenerics_0.36.1 MatrixGenerics_1.2.1
[15] matrixStats_0.58.0 magrittr_2.0.1
[17] workflowr_1.6.2
loaded via a namespace (and not attached):
[1] bitops_1.0-6 fs_1.5.0
[3] httr_1.4.2 rprojroot_2.0.2
[5] tools_4.0.3 R6_2.5.0
[7] irlba_2.3.3 vipor_0.4.5
[9] DBI_1.1.1 lazyeval_0.2.2
[11] colorspace_2.0-0 withr_2.4.1
[13] tidyselect_1.1.0 gridExtra_2.3
[15] compiler_4.0.3 git2r_0.28.0
[17] cli_2.3.0 BiocNeighbors_1.8.2
[19] DelayedArray_0.16.3 plotly_4.9.3
[21] labeling_0.4.2 scales_1.1.1
[23] systemfonts_1.0.1 stringr_1.4.0
[25] digest_0.6.27 rmarkdown_2.6
[27] svglite_1.2.3.2 XVector_0.30.0
[29] RhpcBLASctl_0.20-137 scater_1.18.6
[31] pkgconfig_2.0.3 htmltools_0.5.1.1
[33] sparseMatrixStats_1.2.1 highr_0.8
[35] htmlwidgets_1.5.3 rlang_0.4.10
[37] DelayedMatrixStats_1.12.3 generics_0.1.0
[39] farver_2.0.3 jsonlite_1.7.2
[41] dplyr_1.0.4 RCurl_1.98-1.2
[43] GenomeInfoDbData_1.2.4 scuttle_1.0.4
[45] Matrix_1.3-2 Rcpp_1.0.6
[47] ggbeeswarm_0.6.0 munsell_0.5.0
[49] fansi_0.4.2 viridis_0.5.1
[51] lifecycle_1.0.0 stringi_1.5.3
[53] whisker_0.4 yaml_2.2.1
[55] zlibbioc_1.36.0 grid_4.0.3
[57] promises_1.2.0.1 crayon_1.4.1
[59] lattice_0.20-41 cowplot_1.1.1
[61] beachmat_2.6.4 knitr_1.31
[63] pillar_1.4.7 glue_1.4.2
[65] evaluate_0.14 data.table_1.13.6
[67] vctrs_0.3.6 httpuv_1.5.5
[69] gtable_0.3.0 purrr_0.3.4
[71] tidyr_1.1.2 assertthat_0.2.1
[73] xfun_0.21 rsvd_1.0.3
[75] later_1.1.0.1 viridisLite_0.3.0
[77] tibble_3.0.6 beeswarm_0.2.3
[79] ellipsis_0.3.1 here_1.0.1