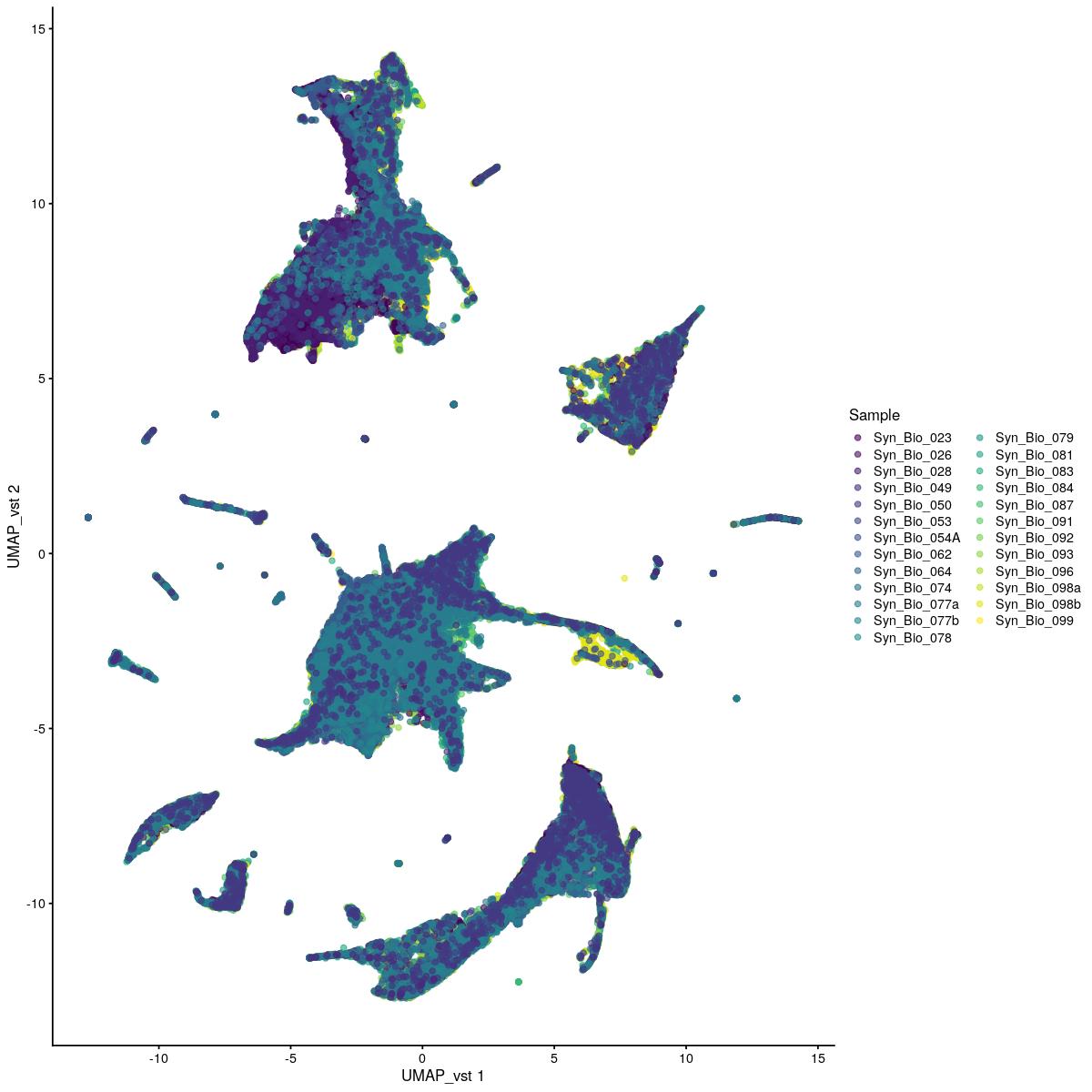
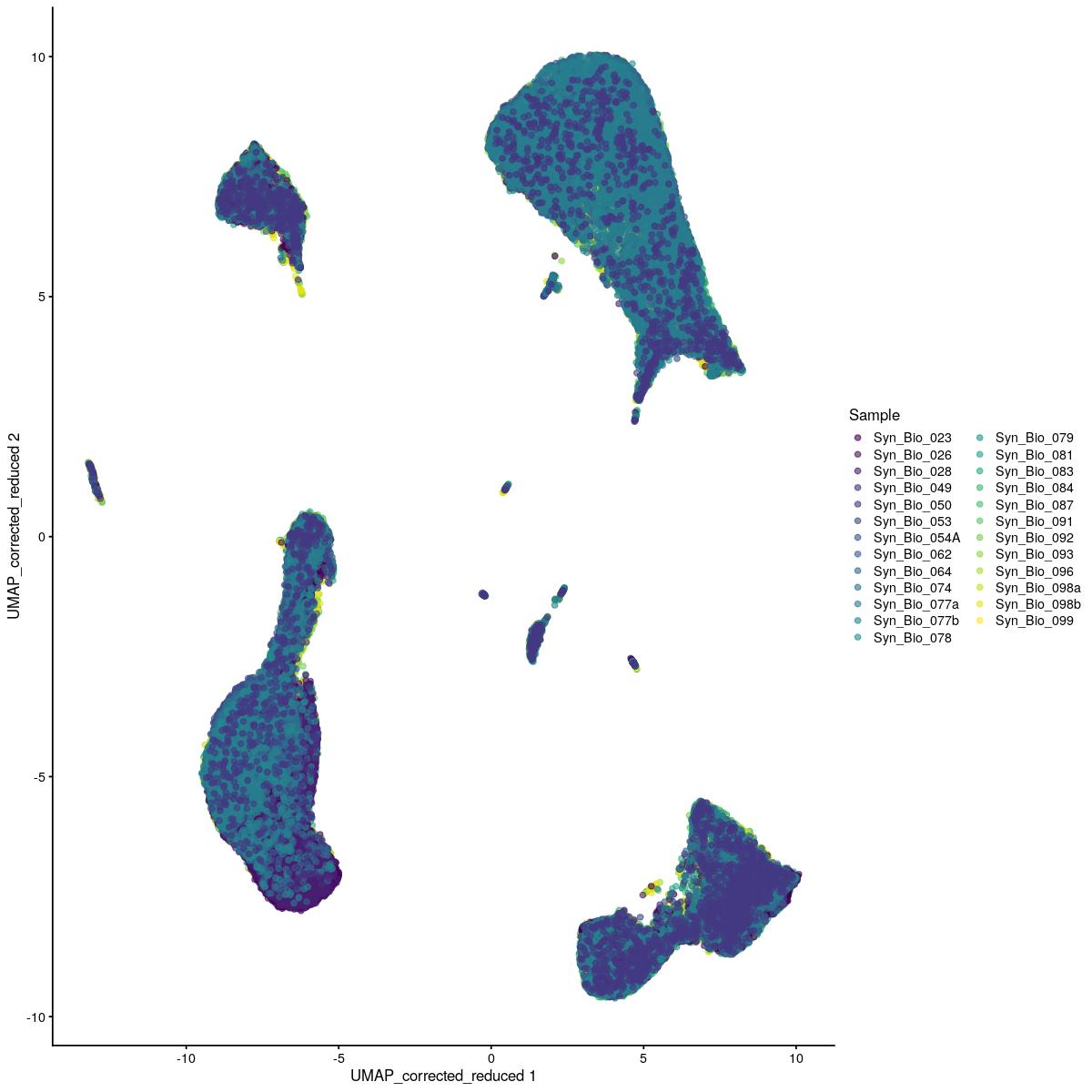
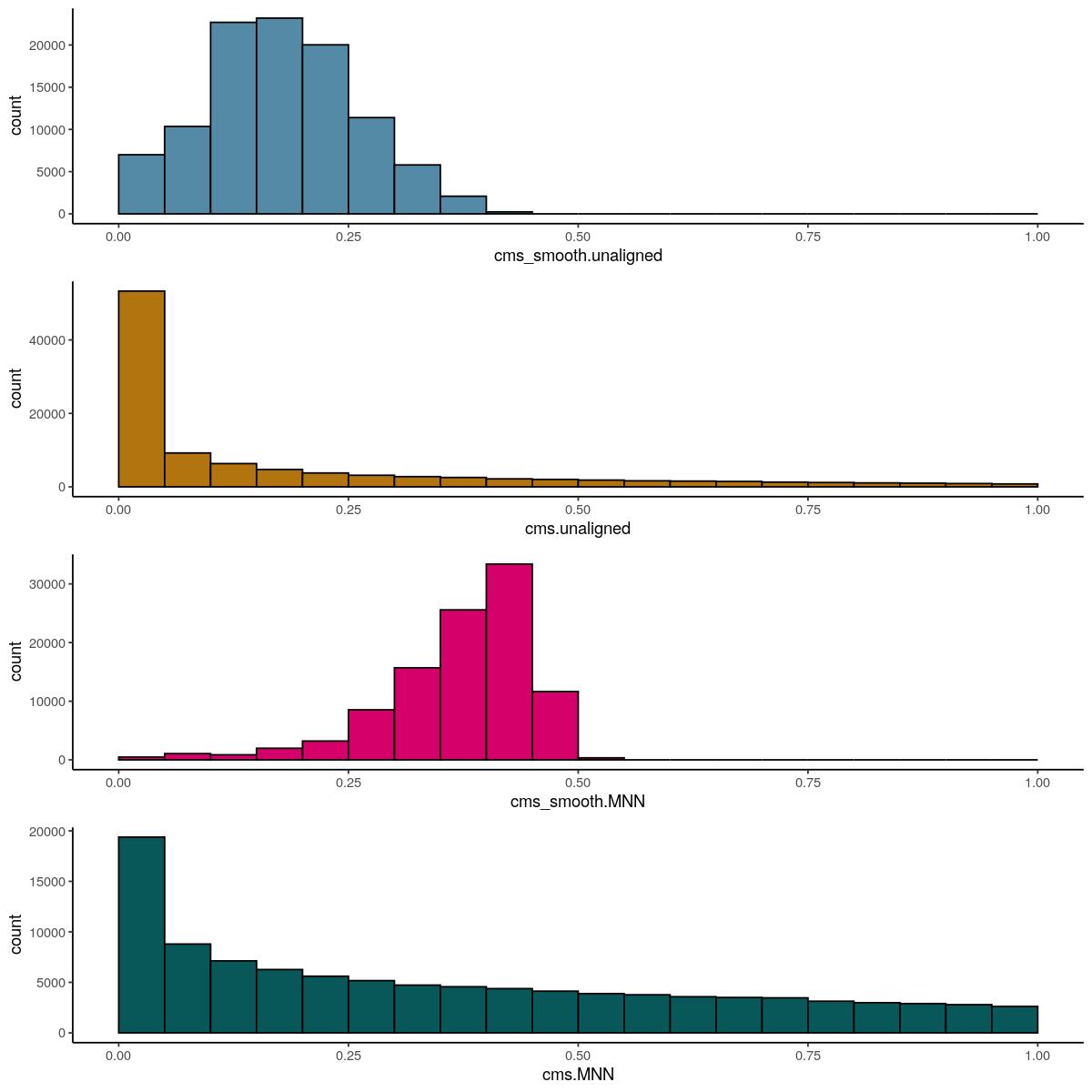
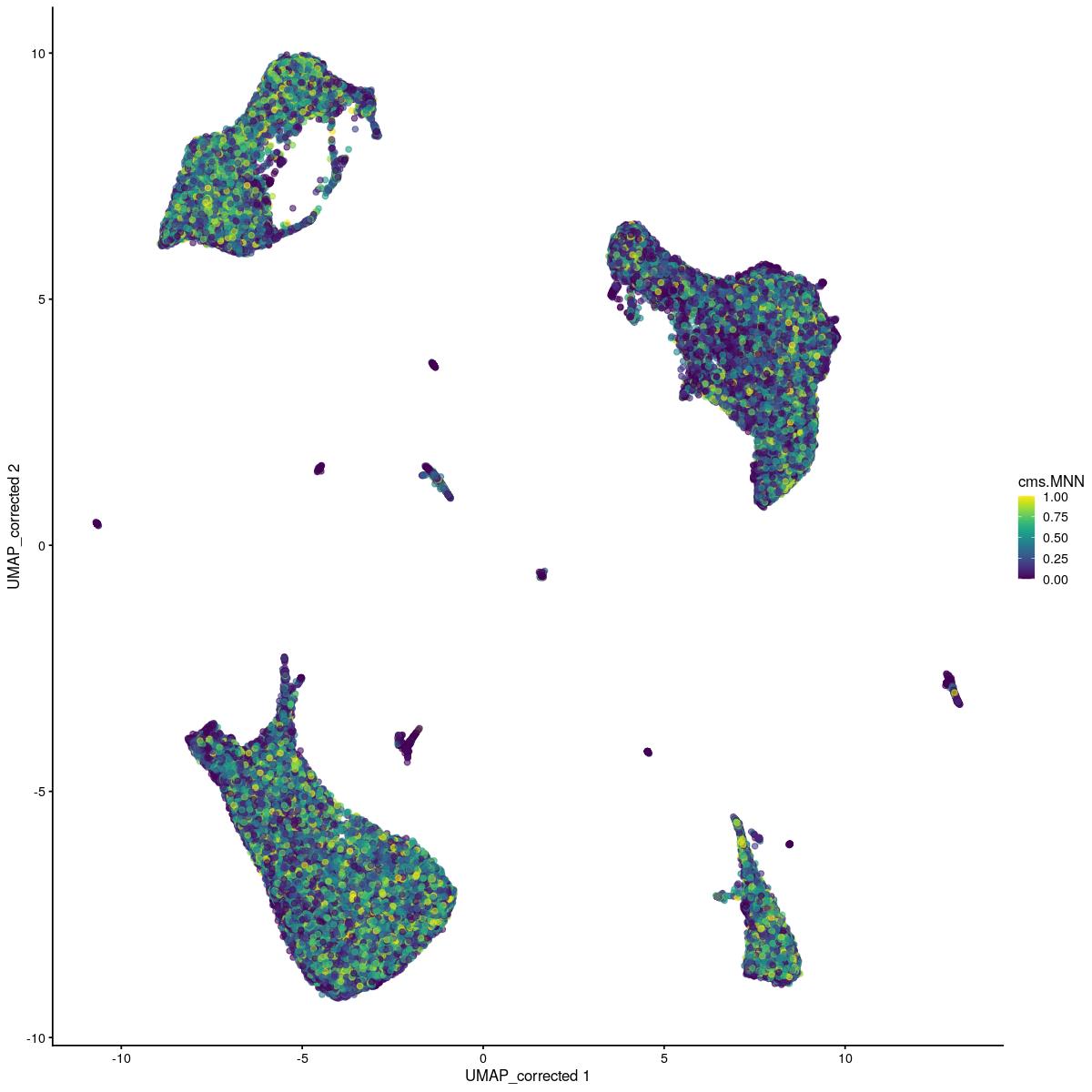
Main Celltype
panglao <- readr::read_tsv("https://panglaodb.se/markers/PanglaoDB_markers_27_Mar_2020.tsv.gz")
── Column specification ────────────────────────────────────────────────────────
cols(
species = col_character(),
`official gene symbol` = col_character(),
`cell type` = col_character(),
nicknames = col_character(),
`ubiquitousness index` = col_double(),
`product description` = col_character(),
`gene type` = col_character(),
`canonical marker` = col_double(),
`germ layer` = col_character(),
organ = col_character(),
sensitivity_human = col_double(),
sensitivity_mouse = col_double(),
specificity_human = col_double(),
specificity_mouse = col_double()
)panglao$organ %>% unique [1] "Pancreas" "Connective tissue" "Brain"
[4] "Lungs" "Smooth muscle" "Immune system"
[7] "Epithelium" "Heart" "Liver"
[10] "Adrenal glands" "GI tract" "Reproductive"
[13] "Kidney" "Zygote" "Vasculature"
[16] "Embryo" "Blood" "Thyroid"
[19] NA "Bone" "Skin"
[22] "Mammary gland" "Eye" "Skeletal muscle"
[25] "Olfactory system" "Parathyroid glands" "Oral cavity"
[28] "Thymus" "Placenta" "Urinary bladder" # restricting the analysis to pancreas specific gene signatues
panglao_sub <- panglao %>% filter(organ %in% c("Connective tissue","Epithelium","Blood","Immune system","Vasculature"))
# restricting to human specific genes
panglao_sub <- panglao_sub %>% filter(str_detect(species,"Hs"))
panglao_sub <- panglao_sub %>% filter(`cell type` %in% c("Dendritic cells","B cells","Fibroblasts","Macrophages","Monocytes","Mast cells","Neutrophils","NK cells","T cells","Endothelial cells", "Neutrophils","Pericytes"))
panglao_sub <- panglao_sub %>%
group_by(`cell type`) %>%
summarise(geneset = list(`official gene symbol`))
pancreas_gs <- setNames(panglao_sub$geneset, panglao_sub$`cell type`)
print(pancreas_gs)$`B cells`
[1] "CD2" "CD5" "MS4A1" "CR2" "CD22" "FCER2"
[7] "CD40" "CD69" "CD70" "CD79A" "CD79B" "CD80"
[13] "CD86" "TNFRSF9" "SDC1" "TNFSF4" "TNFRSF13B" "TNFRSF13C"
[19] "PDCD1" "IGHD" "IGHM" "RASGRP3" "HLA-DRA" "LTB"
[25] "HLA-DQA1" "FLI1" "CD14" "SEMA6D" "LAIR1" "IFIT3"
[31] "IGLL1" "DNTT" "MME" "SPN" "CD19" "CD24"
[37] "CD27" "B3GAT1" "CD72" "MUM1" "PAX5" "JCHAIN"
[43] "MZB1" "LY6D" "FCMR" "BANK1" "EDEM1" "VPREB3"
[49] "POU2AF1" "CRELD2" "DERL3" "RALGPS2" "FCHSD2" "POLD4"
[55] "TNFRSF17" "HVCN1" "FCRLA" "EDEM2" "BLNK" "TXNDC11"
[61] "BTLA" "SMAP2" "FKBP11" "SEC61A1" "SPCS3" "SPIB"
[67] "EAF2" "CXCR4" "BIRC3" "IGLC2" "IGLC3" "IGLC1"
[73] "IL21R" "IGKC" "VPREB1" "LRMP" "KLHL6" "SLAMF6"
[79] "FAM129C" "BST1" "MSH5" "DOK3" "BACH2" "PXK"
[85] "IGHG1" "IGHG3" "IGHG4" "CD38" "PTPRC" "EBF1"
[91] "BCL11A" "CCR7" "CD55" "CD74" "CD52" "TLR9"
[97] "SWAP70" "HMGA1"
$`Dendritic cells`
[1] "IL6" "CD86" "CD83" "CD1A" "CR2" "TLR9"
[7] "CD1C" "CD209" "LAMP3" "CD1B" "TREM2" "FABP4"
[13] "S100A9" "ARG1" "HLA-DRA" "HLA-DQA1" "HLA-DMB" "HLA-DMA"
[19] "HLA-DQB1" "CLEC10A" "HLA-DRB1" "HLA-DPA1" "HLA-DPB1" "DNASE1L3"
[25] "CLEC9A" "LILRB2" "ETV6" "CD163" "CXCR4" "CXCL8"
[31] "VSIG4" "NR4A3" "CCR7" "TRAF1" "RELB" "BATF3"
[37] "CCL22" "SLAMF7" "XCR1" "CXCL16" "SCIMP" "FCGR2B"
[43] "FGD2" "RAB7B" "NAAA" "HCK" "CD180" "HFE"
[49] "CCR2" "RYR1" "ITGAE" "SEMA4A" "DPP4" "SLAMF8"
[55] "CXCR3" "BTLA" "FLT3" "TLR3" "ITGAX" "GPR132"
[61] "ADAM19" "AP1S3" "ASS1" "ADGRG5" "GPR68" "KIT"
[67] "KMO" "P2RY10" "RAB30" "SEPT6" "ZBTB46" "S100A4"
[73] "CLEC7A" "AIF1" "LST1" "CTSS" "IRF8" "ADGRE1"
[79] "CCL17" "CD14" "CD207" "CD8A" "CX3CR1" "ITGAM"
[85] "LY75" "PDCD1LG2" "PTPRC" "SIRPA" "FCGR3A" "FTL"
[91] "SERPINA1" "AXL" "PPP1R14A" "SIGLEC6" "CD22" "DAB2"
[97] "S100A8" "VCAN" "LYZ" "ANXA1" "FCER1A" "C1ORF54"
[103] "CADM1" "CAMK2D" "LGALS3" "NAPSA" "PLBD1" "RNASE6"
[109] "PLAC8" "H2AFY" "SLC11A1" "PDPN" "S100B" "CD28"
[115] "PPL" "SLURP1" "HLA-A" "HLA-B" "HLA-C" "HLA-DRB5"
[121] "SERPINB9"
$`Endothelial cells`
[1] "PECAM1" "ICAM1" "ITGB3" "SELE" "VCAM1" "MCAM"
[7] "PROCR" "TEK" "FLT4" "APLN" "VWF" "NOS3"
[13] "THBD" "PLVAP" "ACKR1" "SLCO1C1" "TMEM100" "ADGRF5"
[19] "ABCG2" "PODXL" "NOSTRIN" "MFSD2A" "ACVRL1" "AQP1"
[25] "MYLK" "RASIP1" "FLI1" "TIE1" "APLNR" "NRP2"
[31] "ADAMTS1" "RPRM" "FABP4" "GPIHBP1" "FHL2" "LOX"
[37] "KLK1" "ADORA2A" "ARAP3" "ARHGEF15" "CARD10" "CLEC14A"
[43] "DLL4" "ESM1" "GIMAP5" "GJA4" "MMRN2" "NOTCH4"
[49] "NPR1" "PRKCH" "RASGRP3" "ROBO4" "SCARF1" "SOX18"
[55] "SOX7" "SPNS2" "THSD1" "APOLD1" "EMP1" "CD36"
[61] "RNASE1" "CTGF" "HYAL2" "CLEC4G" "GPR182" "F8"
[67] "RBP7" "CALCRL" "FOXF1" "CASZ1" "AQP7" "TCF15"
[73] "CD300LG" "BTNL9" "MEOX2" "ERG" "HEXIM1" "GLYCAM1"
[79] "CD55" "MMRN1" "C7" "RAMP3" "VEGFC" "GJA5"
[85] "HEY1" "RND1" "BDP1" "CD46" "MEOX1" "CCL19"
[91] "MADCAM1" "CYP1B1" "IRX3" "BIRC2" "LYVE1" "SEMA3D"
[97] "EMCN" "WFDC1" "ADGRL4" "VWA1" "ECE1" "PTPRB"
[103] "CLDN5" "TBX1" "SEMA7A" "FOXF2" "PDGFB" "ECSCR"
[109] "ELK3" "CDH5" "PLEC" "STAB1" "TGFBR2" "CD93"
[115] "CXCL1" "RGS5" "SLC7A5" "ENG" "KDR" "SLC2A1"
[121] "EGFL7" "FLT1" "EPAS1" "EDNRB" "KCNJ8" "CD82"
[127] "CHST1" "PLAC8" "TSPAN8" "ETS1" "CD34" "PDPN"
[133] "PROX1" "EHD3" "SRGN" "S100A10" "CLIC4" "USHBP1"
[139] "MYF6" "OIT3" "IL1A" "BMP2" "C1QTNF1" "PCDH12"
[145] "DPP4" "IGFBP7" "PALMD" "POSTN" "BMX" "SLC38A5"
[151] "XDH" "SPARC" "MGLL" "SLC9A3R2" "RGCC" "ICAM2"
[157] "MGP" "SPARCL1" "TM4SF1" "ID1" "ADIRF" "CD9"
[163] "SRPX" "ID3" "CAV1" "GNG11" "HSPG2" "CCL14"
[169] "CLEC1B" "FCN2" "S100A13" "FCN3" "CRHBP" "IFI27"
[175] "CCL23" "SGK1" "DNASE1L3" "LIFR" "PCAT19" "CDKN1C"
[181] "INMT" "PTGDS" "TIMP3" "GPM6A" "FAM167B" "LTC4S"
[187] "STAB2"
$Fibroblasts
[1] "IL1R1" "FAP" "FLI1" "CELA1" "LOX" "PDGFRB"
[7] "P4HA1" "UCP2" "CCR2" "ITGAL" "FGR" "HCK"
[13] "TNFRSF1B" "PRKCD" "ENO3" "ABI3" "TREML4" "PIP4K2A"
[19] "CD300E" "SERPINB10" "CTHRC1" "TBX18" "COL15A1" "GJB2"
[25] "IL34" "EDN3" "SLC6A13" "VTN" "ITIH5" "LUM"
[31] "DPT" "POSTN" "PENK" "MMP14" "COL6A2" "FABP4"
[37] "ASPN" "ANGPTL2" "EFEMP1" "SCARA5" "IGFBP3" "COPZ2"
[43] "DPEP1" "ADAMTS5" "COL5A1" "CD248" "PI16" "PAMR1"
[49] "TNXB" "MMP2" "COL14A1" "CLEC3B" "IGFBP6" "COL5A2"
[55] "FBN1" "MFAP5" "FKBP10" "PALLD" "WIF1" "SNHG18"
[61] "CDH11" "PTCH1" "ARAP1" "FBLN2" "IGF1" "PRRX1"
[67] "FKBP7" "OAF" "COL6A3" "CTSK" "DKK1" "C1S"
[73] "RARRES2" "GREM1" "SPON2" "TCF21" "PCSK6" "COL8A1"
[79] "ENTPD2" "CXCL8" "CXCL3" "IL6" "CYP1B1" "COL13A1"
[85] "ADAMTS10" "CCL11" "ADAM33" "COL4A3" "COL4A4" "LAMA2"
[91] "ACKR3" "CD55" "FBLN7" "FIBIN" "THBS2" "NOV"
[97] "PTX3" "MMP3" "LRRK1" "HGF" "FRZB" "COL12A1"
[103] "COL7A1" "MEOX1" "PRG4" "PKD2" "CCL19" "NNMT"
[109] "FOXF1" "HAS1" "CTGF" "ERCC1" "WISP1" "TWIST2"
[115] "RIPK3" "DDR2" "ELN" "FN1" "HHIP" "FMO2"
[121] "COL1A2" "COL3A1" "VIM" "FSTL1" "GSN" "SPARC"
[127] "S100A4" "NT5E" "COL1A1" "MGP" "NOX4" "THY1"
[133] "CD40" "SERPINH1" "CD44" "PDGFRA" "EN1" "DCN"
[139] "CEBPB" "EGR1" "FOSB" "FOSL2" "HIF1A" "KLF2"
[145] "KLF4" "KLF6" "KLF9" "NFAT5" "NFATC1" "NFKB1"
[151] "NR4A1" "NR4A2" "PBX1" "RUNX1" "STAT3" "TCF4"
[157] "ZEB2" "LAMC1" "MEDAG" "LAMB1" "DKK3" "TBX20"
[163] "MDK" "GSTM5" "NGF" "VEGFA" "FGF2" "P4HTM"
[169] "CKAP4" "INMT" "CXCL14"
$Macrophages
[1] "ITGAL" "ITGAM" "CD14" "FUT4" "FCGR3A" "CD33"
[7] "FCGR1A" "CD80" "LILRB4" "CD86" "CD163" "CCR5"
[13] "TLR2" "TLR4" "ADGRE1" "GPR34" "TREM2" "FABP4"
[19] "S100A8" "CPM" "CHIT1" "F13A1" "CX3CR1" "CXCL16"
[25] "TGFBR1" "SLAMF9" "SCIMP" "LILRA5" "CD83" "C3AR1"
[31] "STAB1" "MRC1" "PARP14" "FGD2" "RAB7B" "RBPJ"
[37] "SLCO2B1" "NAAA" "MARCH1" "EGLN3" "JAML" "FGL2"
[43] "GPNMB" "CLEC4D" "ADAM8" "ARL11" "MMP12" "LYVE1"
[49] "PLTP" "VSIG4" "MS4A4A" "MS4A6A" "FPR1" "CD180"
[55] "GDF15" "RAB20" "HFE" "TNF" "CCR2" "SNX20"
[61] "FMNL1" "GPR132" "SLAMF7" "NCEH1" "CCL24" "C5AR1"
[67] "CD300A" "CXCL2" "CCL7" "CCL2" "IL1B" "IRF5"
[73] "AHR" "MYO1G" "DUSP5" "GPR171" "CCR7" "DNASE1L3"
[79] "CXCL1" "SAMSN1" "NR4A3" "CCL22" "S100A4" "MMP9"
[85] "HILPDA" "NRP2" "SLC37A2" "CTSK" "CD36" "LPCAT2"
[91] "HPGDS" "IFNAR2" "MS4A7" "SLC11A1" "HPGD" "CCL3"
[97] "CLEC7A" "CD5L" "CCL5" "CYTH4" "CD3E" "CD19"
[103] "CD74" "CSF1R" "LGALS3" "CD68" "UCP2" "TREML4"
[109] "FGR" "CYBB" "CD200" "CD200R1" "GATA6" "ITGAX"
[115] "PPARG" "TYROBP" "RGS1" "DAB2" "P2RY6" "MAF"
[121] "AIF1" "CLEC10A" "ADGRE5" "SLC15A3" "CYP27A1" "SLC7A7"
[127] "RUNX3" "SYK"
$`Mast cells`
[1] "KIT" "ENPP3" "IL2RA" "FCER2" "IL17A"
[6] "CRH" "HSD11B1" "CD274" "CXCR2" "CXCR4"
[11] "CCR3" "CCR5" "LTC4S" "GPR34" "MGST2"
[16] "ACHE" "CPA3" "CPA1" "CMA1" "HNMT"
[21] "TPSAB1" "PTGDS" "CFP" "RAB27B" "SLA"
[26] "IL1RL1" "GNAI1" "CFD" "OSBPL8" "ADORA3"
[31] "HS3ST1" "CSF2RB" "DAPP1" "SLC6A4" "MAOB"
[36] "CSRNP1" "CYP11A1" "RGS1" "PLEK" "HDC"
[41] "TPSB2" "TPH1" "CD55" "RGS13" "TPSG1"
[46] "SOCS1" "BTK" "IL4" "MS4A2" "VWA5A"
[51] "FCER1A" "MCEMP1" "MILR1" "CCL2" "CREB3L1"
[56] "EDNRA" "HS6ST2" "KCNE3" "MEIS2" "MRGPRX2"
[61] "PLAU" "ADAMTS9" "CHST1" "COBL" "CTSG"
[66] "DNM3" "FAM198B" "GRIK2" "HPGDS" "HS3ST3A1"
[71] "PAPSS2" "PCBD1" "PCP4L1" "RNF128" "SLC29A1"
[76] "SLC45A3" "STARD13" "A4GALT" "ACER3" "ARMCX3"
[81] "ATL2" "BBS10" "BMPR2" "C2" "CCDC141"
[86] "CCL7" "CDC42BPA" "CDH9" "COPZ2" "ZDHHC13"
[91] "WDR60" "UNC13B" "TRANK1" "TIAM2" "TCF7L1"
[96] "STK32B" "ST6GALNAC3" "SOCS2" "SPRED1" "SMPX"
[101] "SMARCA1" "SLC7A5" "SLC4A4" "SLC31A2" "SGCE"
[106] "ROR1" "RNF180" "RIOK2" "RBFOX1" "RAPGEF2"
[111] "PDE1C" "PCDH7" "OPTN" "OAF" "NEO1"
[116] "NDST2" "NCEH1" "MRGPRX1" "MLPH" "MITF"
[121] "MFGE8" "LSAMP" "LRRC66" "LIMA1" "KRT4"
[126] "KDELR3" "IDS" "HSPA13" "HS2ST1" "GPM6A"
[131] "GP1BA" "GNAZ" "FAM84A" "FAM129B" "EXT1"
[136] "ESYT3" "ENO2" "ENAH" "DUSP1" "DGKI"
[141] "DDC" "DCLK3"
$Monocytes
[1] "RGS1" "APOBEC3A" "CD7" "TET2" "CD40" "DYSF"
[7] "CMKLR1" "MEFV" "HCK" "FCGR3B" "PADI4" "GHSR"
[13] "ITGAX" "SELE" "TLR4" "AR" "CXCR4" "CD86"
[19] "CCR2" "TNFRSF14" "ADA2" "MGMT" "CD14" "CD33"
[25] "ITGAM" "ACE" "FUT4" "ICAM1" "SELL" "CD163"
[31] "FCGR1A" "FCGR2B" "ACP5" "MRC1" "TNF" "PLAU"
[37] "GBP1" "OAS1" "IRF7" "PLSCR1" "MX1" "IL1RN"
[43] "HLA-DRA" "IFIT1" "IDO1" "IFIT3" "TNFSF10" "CXCL10"
[49] "S100A9" "S100A8" "S100A4" "CLEC7A" "CSF3R" "MNDA"
[55] "MS4A6A" "ZFP36L2" "LTA4H" "CLEC12A" "CD48" "PRTN3"
[61] "FCGR3A" "VCAN" "IFITM3" "FN1" "ADGRE1" "CD44"
[67] "CSF1R" "CX3CR1" "ITGAL" "PECAM1" "PTPRC" "SPN"
[73] "PSAP" "FCN1" "LYZ" "RHOC" "PILRA" "NFKBIZ"
[79] "NAAA" "LY6E" "LYN" "MS4A7" "CEBPB" "CD68"
[85] "IFI30" "S100A12" "SERPINA1" "RGS2" "LST1" "SPI1"
[91] "TYMP" "CSTA" "FGL2" "PYCARD" "LYST" "CCL3"
[97] "IL1B" "CFP" "CD36"
$Neutrophils
[1] "MME" "ITGAM" "ITGAX" "CD14" "FUT4" "FCGR3A"
[7] "PECAM1" "CD33" "SELL" "CEACAM8" "C5AR1" "CXCR1"
[13] "CXCR2" "JAML" "TLR2" "MYLK" "S100A9" "MPO"
[19] "CD24" "CEACAM1" "FCGR1A" "CRP" "LCN2" "DEFA1"
[25] "DEFA3" "LTF" "LYZ" "ELANE" "S100A8" "IL1B"
[31] "CXCL2" "HP" "CCL3" "HDC" "MMP8" "LRG1"
[37] "OSM" "MMP9" "PILRA" "CLEC4D" "CLEC4E" "ASPRV1"
[43] "CCRL2" "CCR1" "NCF1" "TREM1" "SORL1" "ARG2"
[49] "BST1" "IL1R2" "CFP" "ADAM8" "CD177" "PTGS2"
[55] "OAS3" "PRTN3" "AZU1" "CTSG" "SERPINB1" "CAMP"
[61] "SLC1A5" "SNX20" "ADPGK" "PSTPIP1" "LYST" "DOCK8"
[67] "S100A4" "NLRP3" "CSF3R"
$`NK cells`
[1] "ITGAM" "ITGAX" "FCGR3A" "CD69" "KLRD1" "IL2RB"
[7] "KLRB1" "CD244" "KLRK1" "SLAMF7" "SIGLEC7" "NCR1"
[13] "SLAMF6" "KIT" "CD27" "KLRC1" "KLRF1" "GNLY"
[19] "NKG7" "IL32" "GZMH" "FGFBP2" "GZMM" "CTSW"
[25] "HMBOX1" "AHR" "PRF1" "CCL4" "SEMA6D" "FHL2"
[31] "CD2" "CD7" "CD3G" "CD33" "DPP4" "LAT"
[37] "PCDH15" "CCL5" "GZMA" "GZMB" "CMA1" "DUSP2"
[43] "TXK" "DOK2" "CST7" "HSD11B1" "KLRC2" "SAMD3"
[49] "TBX21" "CHSY1" "XCL2" "TRDC" "CXCR4" "IL18R1"
[55] "SERPINB9" "DOCK2" "SH2D2A" "S100A4" "XCL1" "GZMK"
[61] "IFNG" "CCL3" "CSF2" "IL2RG" "TGFB1" "KIR2DL1"
[67] "KIR3DL1" "LILRB1" "KLRG1" "NCR3" "ADAMTS14" "ITGA2"
[73] "STYK1" "CLEC2D" "CD247" "ZBTB16" "SPON2" "LAIR2"
[79] "HOPX" "CD8A"
$Pericytes
[1] "PECAM1" "PDGFRB" "CSPG4" "ANPEP" "ACTA2" "DES"
[7] "RGS5" "ABCC9" "KCNJ8" "CD248" "DLK1" "TEK"
[13] "NOTCH3" "GLI1" "ICAM1" "ADM" "ANGPT1" "VEGFA"
[19] "ZIC1" "FOXC1" "POSTN" "COX4I2" "HIGD1B" "PDZD2"
[25] "HSD11B1" "MCAM" "MXRA8" "PDE5A" "NR1H3" "SERPING1"
[31] "EMID1" "ECM1" "COLEC11" "RARRES2" "REM1" "ASPN"
[37] "CYGB" "FABP4" "VTN" "STEAP4" "NDUFA4L2" "SLC38A11"
[43] "ATP13A5" "AOC3" "ANGPT2" "INPP4B" "GPIHBP1" "VIM"
[49] "PTH1R" "IFITM1" "TBX18" "NT5E" "MFGE8" "ALPL"
[55] "COL1A1" "MYO1B" "COG7" "P2RY14" "HEYL" "GNB4"
[61] "MSX1" "CTGF"
$`T cells`
[1] "CD3D" "CD3G" "CD3E" "HOPX" "CCL3" "CCL4"
[7] "GIMAP2" "SYT3" "NOTCH3" "SEMA6D" "DKK3" "IFIT3"
[13] "CERK" "PMCH" "CD4" "LTB" "CXCR6" "IL7R"
[19] "SATB1" "LEF1" "ITK" "TRBC2" "PTPRCAP" "GEM"
[25] "CD7" "MAFF" "TGIF1" "RORA" "TNFAIP3" "CREM"
[31] "PXDC1" "NABP1" "FAM110A" "EEF1B2P3" "TRAC" "CD69"
[37] "PFN1P1" "IL32" "CXCR4" "SEPT1" "BCL2" "CYTL1"
[43] "CD2" "CTSW" "PTPN22" "TXK" "GDPD3" "TRAF1"
[49] "IL2RA" "CD8B" "BATF3" "GZMH" "LAG3" "GZMK"
[55] "GZMB" "SH2D1A" "MYO1G" "FMNL1" "S1PR4" "CD247"
[61] "GIMAP5" "CD28" "CD160" "TRDC" "RHOH" "KLRB1"
[67] "CCR2" "IL2RB" "CD163L1" "MBD2" "ICOS" "IL18R1"
[73] "TNFRSF4" "CCL20" "CLEC2D" "CD8A" "CD6" "S100A4"
[79] "CCL5" "LCK" "CD81" "THY1" "LAT" "SKAP1"
[85] "TCF7" "CCR7" "MYB" "CCL4L2" "PYHIN1" "GZMA"
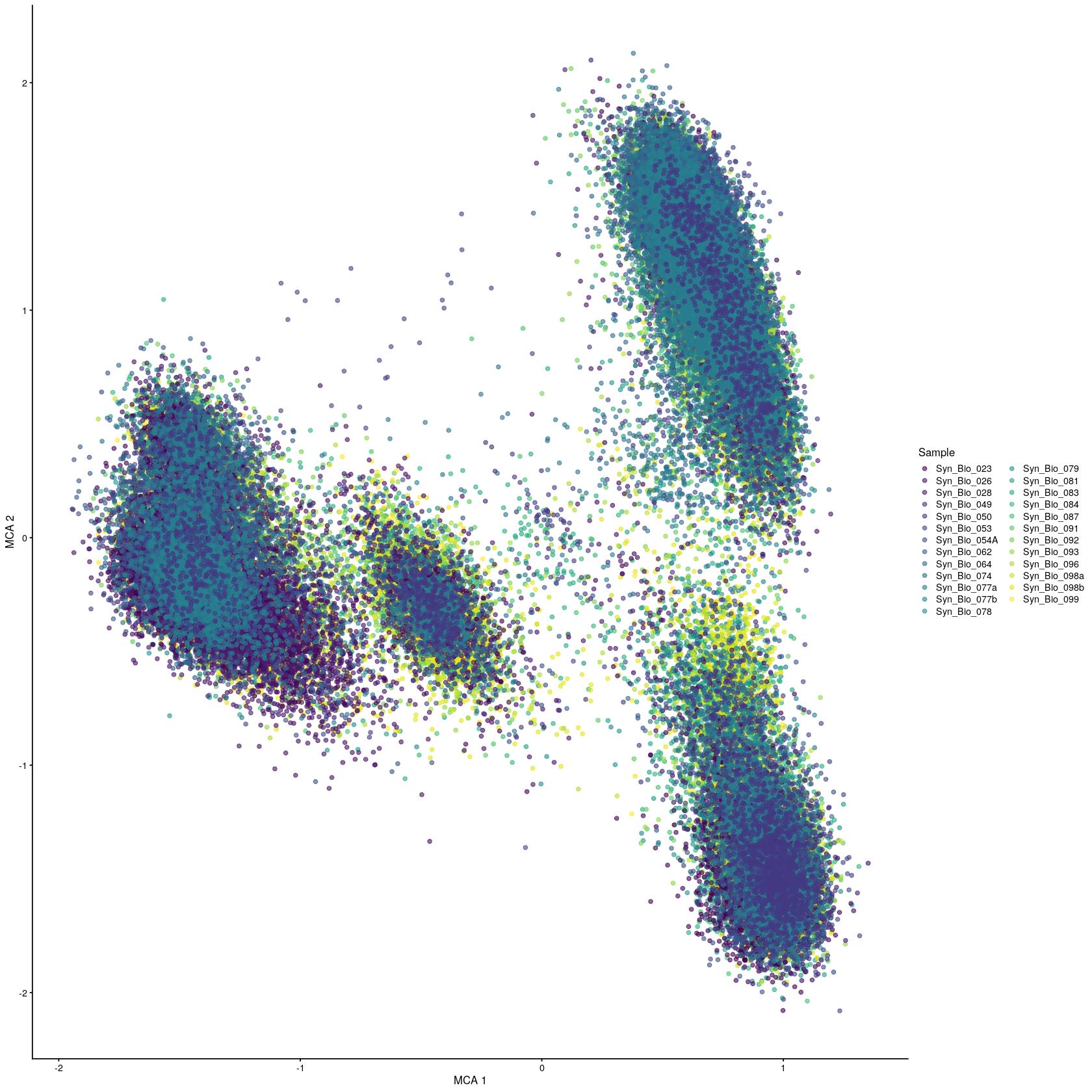
[91] "JUNB" "DUSP2" "IFNG" "CD52" "BRAF" syn_sce_tidy_hvg_cms <- RunMCA(syn_sce_tidy_hvg_cms, slot = "reconstructed")Computing Fuzzy Matrix172.05 sec elapsedComputing SVD198.782 sec elapsedComputing Coordinates33.88 sec elapsedplotReducedDim(syn_sce_tidy_hvg_cms, "MCA", colour_by = "Sample")
Past versions of unnamed-chunk-1-1.jpeg
Version
Author
Date
7d99571
Reto Gerber
2022-03-21
9133ed1
Reto Gerber
2022-03-04
222b0d1
Reto Gerber
2021-07-29
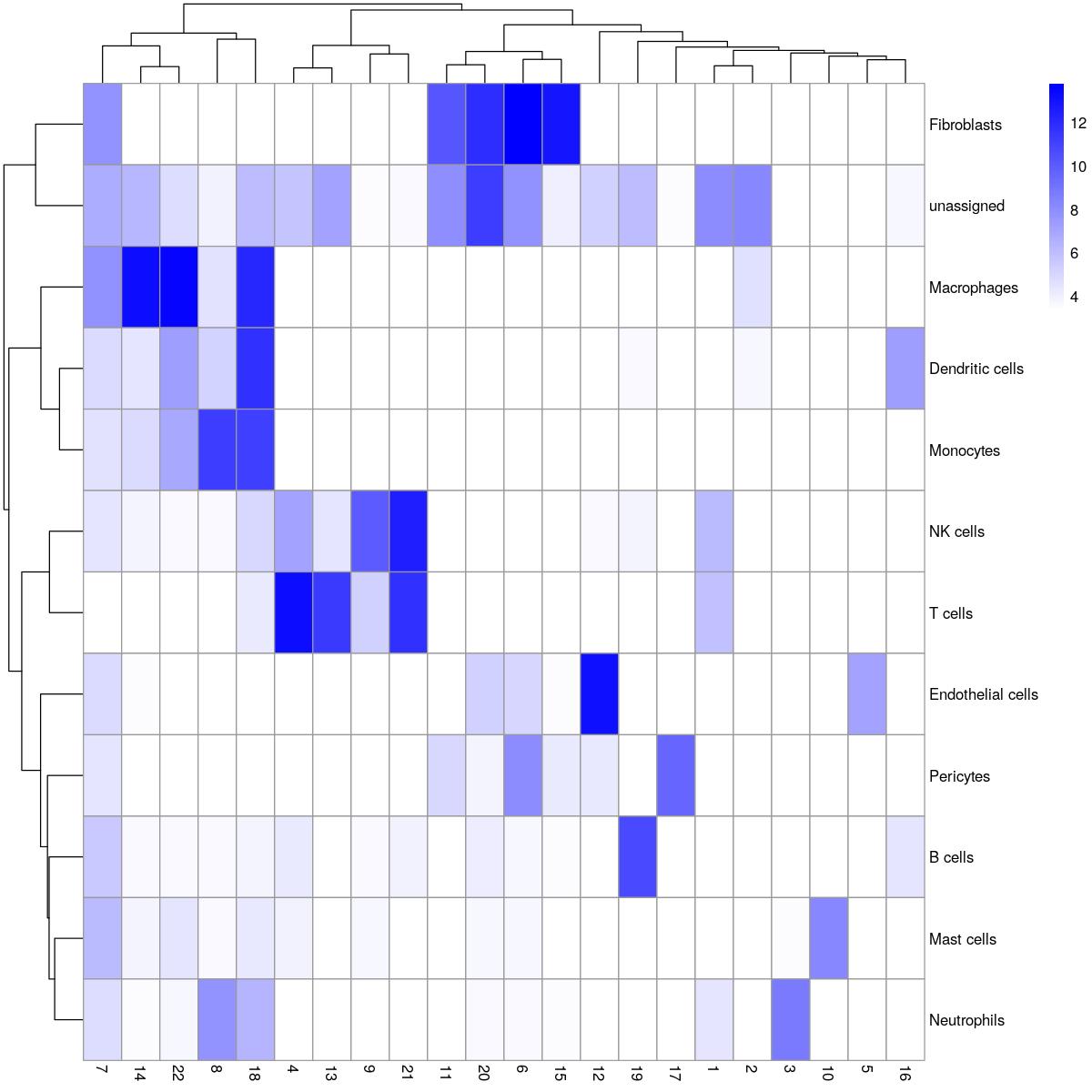
HGT_all_gs <- RunCellHGT(syn_sce_tidy_hvg_cms, pathways = pancreas_gs, minSize = 5)
calculating distanceranking genescalculating number of successperforming hypergeometric test# apply(HGT_all_gs,2,function(i)i[i>2])
all_gs_prediction <- rownames(HGT_all_gs)[apply(HGT_all_gs, 2, which.max)]
all_gs_prediction_signif <- all_gs_prediction
all_gs_prediction_signif <- ifelse(apply(HGT_all_gs, 2, max)>2, yes = all_gs_prediction, "unassigned")
syn_sce_tidy_hvg_cms$main_celltype_cellid <- all_gs_prediction_signif
table(syn_sce_tidy_hvg_cms$main_celltype_cellid, syn_sce_tidy_hvg_cms$Sample)
Syn_Bio_023 Syn_Bio_026 Syn_Bio_028 Syn_Bio_049 Syn_Bio_050
B cells 91 5 1 14 144
Dendritic cells 99 94 124 11 242
Endothelial cells 513 407 148 93 332
Fibroblasts 1989 5995 2784 89 354
Macrophages 819 398 1300 102 431
Mast cells 18 82 3 1 8
Monocytes 308 17 51 11 205
Neutrophils 20 1 1 1 11
NK cells 519 162 70 95 198
Pericytes 27 51 25 6 14
T cells 1197 297 89 84 1075
unassigned 232 334 146 17 137
Syn_Bio_053 Syn_Bio_054A Syn_Bio_062 Syn_Bio_064
B cells 46 40 3 2
Dendritic cells 104 40 156 250
Endothelial cells 29 99 262 92
Fibroblasts 340 616 1166 476
Macrophages 1457 89 2019 2676
Mast cells 18 14 6 6
Monocytes 121 165 30 229
Neutrophils 7 46 2 5
NK cells 481 119 42 125
Pericytes 14 61 25 12
T cells 1099 474 62 315
unassigned 159 197 272 87
Syn_Bio_074 Syn_Bio_077a Syn_Bio_077b Syn_Bio_078
B cells 97 117 78 29
Dendritic cells 153 461 372 3
Endothelial cells 329 150 197 437
Fibroblasts 481 559 885 190
Macrophages 1239 2165 2138 1
Mast cells 47 12 13 3
Monocytes 97 529 597 64
Neutrophils 10 13 28 62
NK cells 203 325 328 185
Pericytes 23 10 15 148
T cells 529 610 482 313
unassigned 194 162 109 11
Syn_Bio_079 Syn_Bio_081 Syn_Bio_083 Syn_Bio_084 Syn_Bio_087
B cells 233 83 375 6 77
Dendritic cells 175 155 164 159 64
Endothelial cells 401 290 162 138 524
Fibroblasts 825 1980 247 530 281
Macrophages 1295 1137 817 1455 278
Mast cells 27 11 9 8 9
Monocytes 130 124 397 402 54
Neutrophils 8 24 68 16 13
NK cells 1333 457 506 165 195
Pericytes 33 34 21 11 33
T cells 1349 739 1361 866 948
unassigned 223 163 132 205 62
Syn_Bio_091 Syn_Bio_092 Syn_Bio_093 Syn_Bio_096
B cells 29 3 3 123
Dendritic cells 153 193 241 145
Endothelial cells 458 802 988 689
Fibroblasts 798 1174 528 1846
Macrophages 1065 2052 1882 600
Mast cells 3 11 1 23
Monocytes 215 169 161 128
Neutrophils 27 6 10 7
NK cells 367 83 156 297
Pericytes 25 32 89 180
T cells 1146 212 211 736
unassigned 90 450 175 202
Syn_Bio_098a Syn_Bio_098b Syn_Bio_099
B cells 111 31 8
Dendritic cells 199 47 94
Endothelial cells 464 556 867
Fibroblasts 240 1556 1371
Macrophages 417 155 436
Mast cells 8 63 7
Monocytes 129 497 118
Neutrophils 10 344 5
NK cells 253 211 42
Pericytes 3 94 95
T cells 1112 462 78
unassigned 39 76 139table(syn_sce_tidy_hvg_cms$main_celltype_cellid)
B cells Dendritic cells Endothelial cells Fibroblasts
1749 3898 9427 27300
Macrophages Mast cells Monocytes Neutrophils
26423 411 4948 745
NK cells Pericytes T cells unassigned
6917 1081 15846 4013 table(syn_sce_tidy_hvg_cms$main_celltype_cellid)/length(syn_sce_tidy_hvg_cms$main_celltype_cellid)
B cells Dendritic cells Endothelial cells Fibroblasts
0.017020573 0.037933786 0.091739816 0.265672746
Macrophages Mast cells Monocytes Neutrophils
0.257138130 0.003999689 0.048151969 0.007250044
NK cells Pericytes T cells unassigned
0.067313494 0.010519862 0.154206972 0.039052920 # colData(syn_sce_tidy_hvg_cms)n_sam <- length(unique(syn_sce_tidy_hvg_cms$main_celltype_cellid))
splitind <- split(seq_len(n_sam),ceiling(seq(0.01,3.99,length.out = n_sam)))
colind <- unlist(purrr::map(seq_len(ceiling(n_sam/4)),
~ purrr::map(seq_len(4),
function(i)splitind[[i]][.x])))
colind <- colind[!is.na(colind)]
colors_used <- rainbow(n_sam)[colind]
cat("### Dimred plots celltype {.tabset}\n\n")
Dimred plots celltype
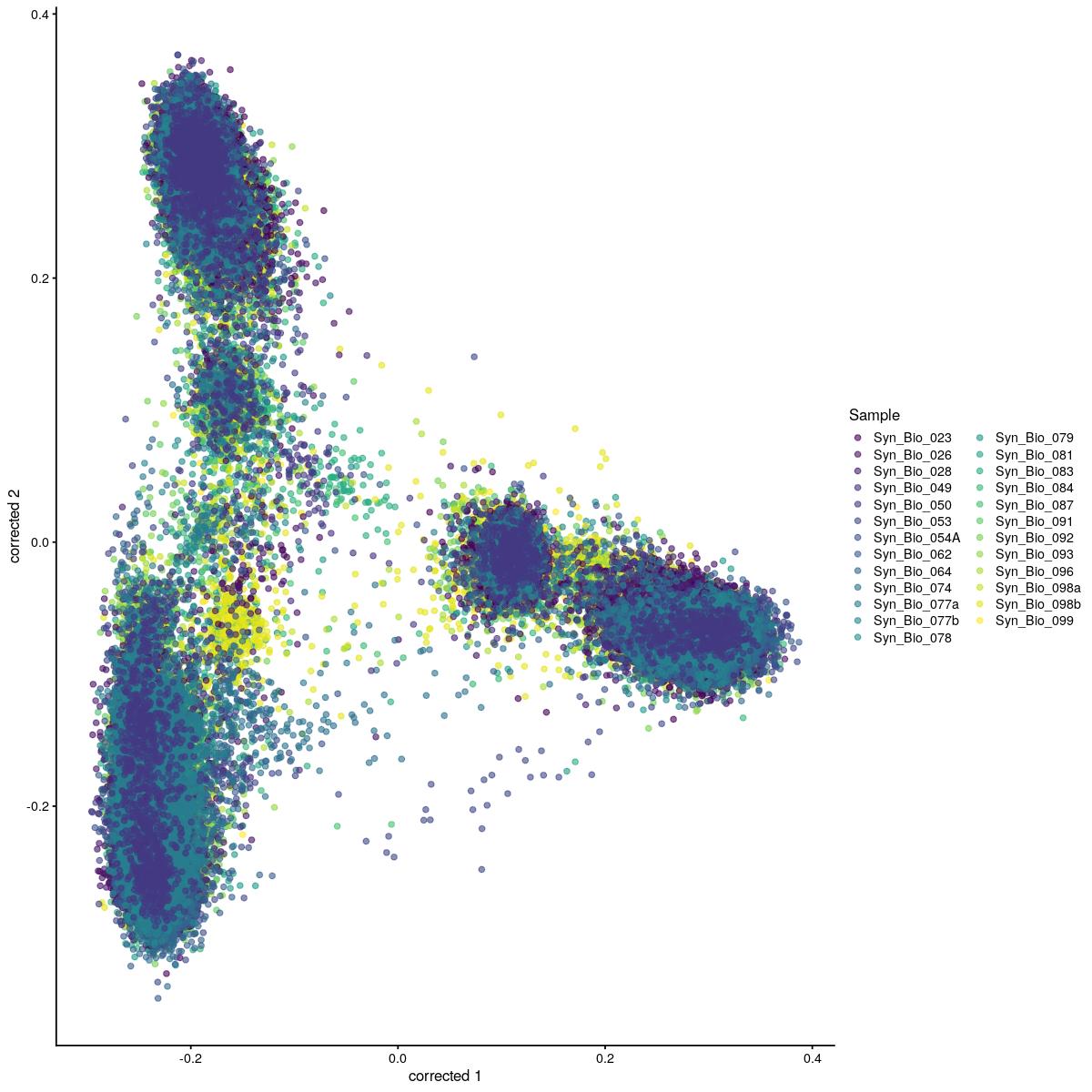
cat("#### corrected PCA\n\n")
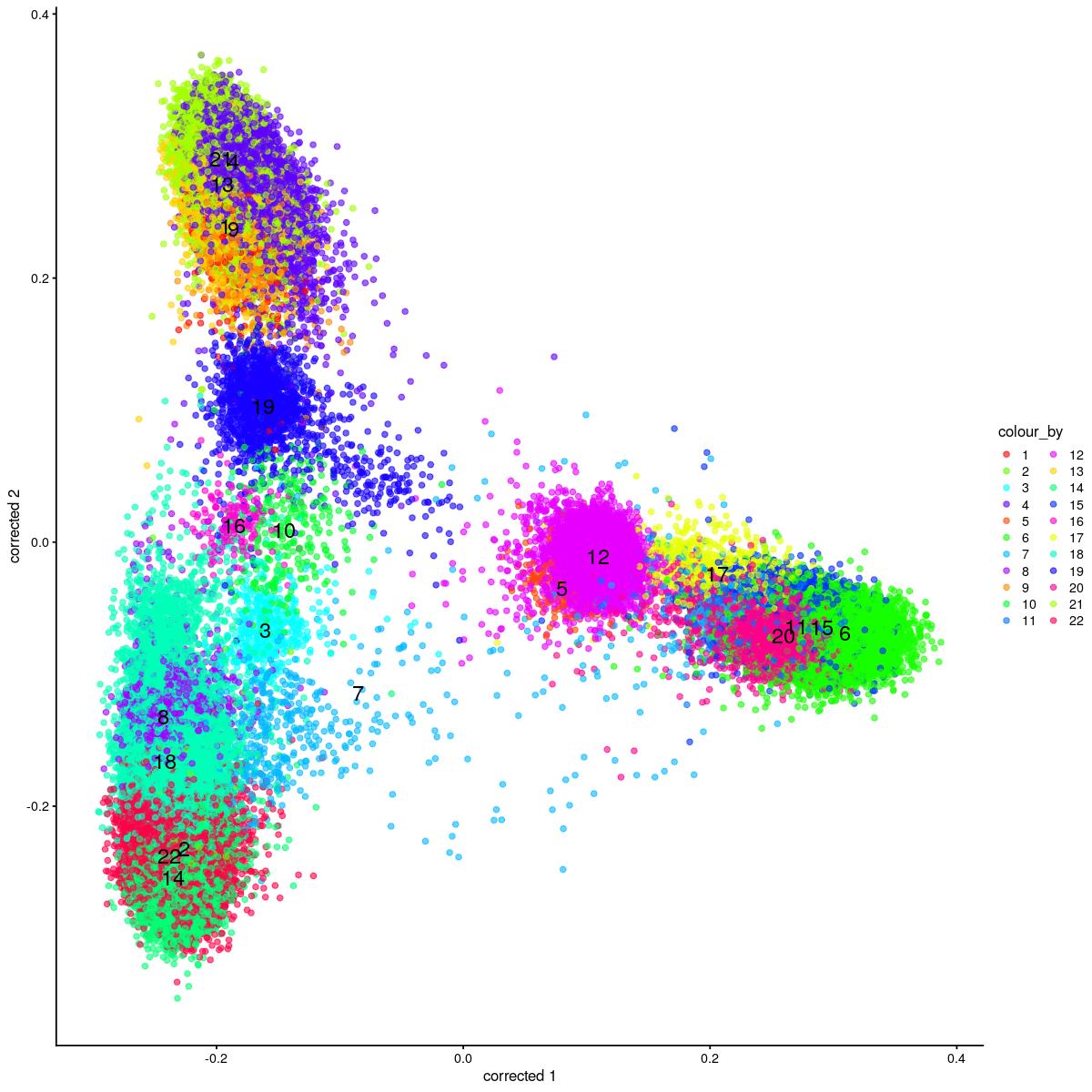
corrected PCA
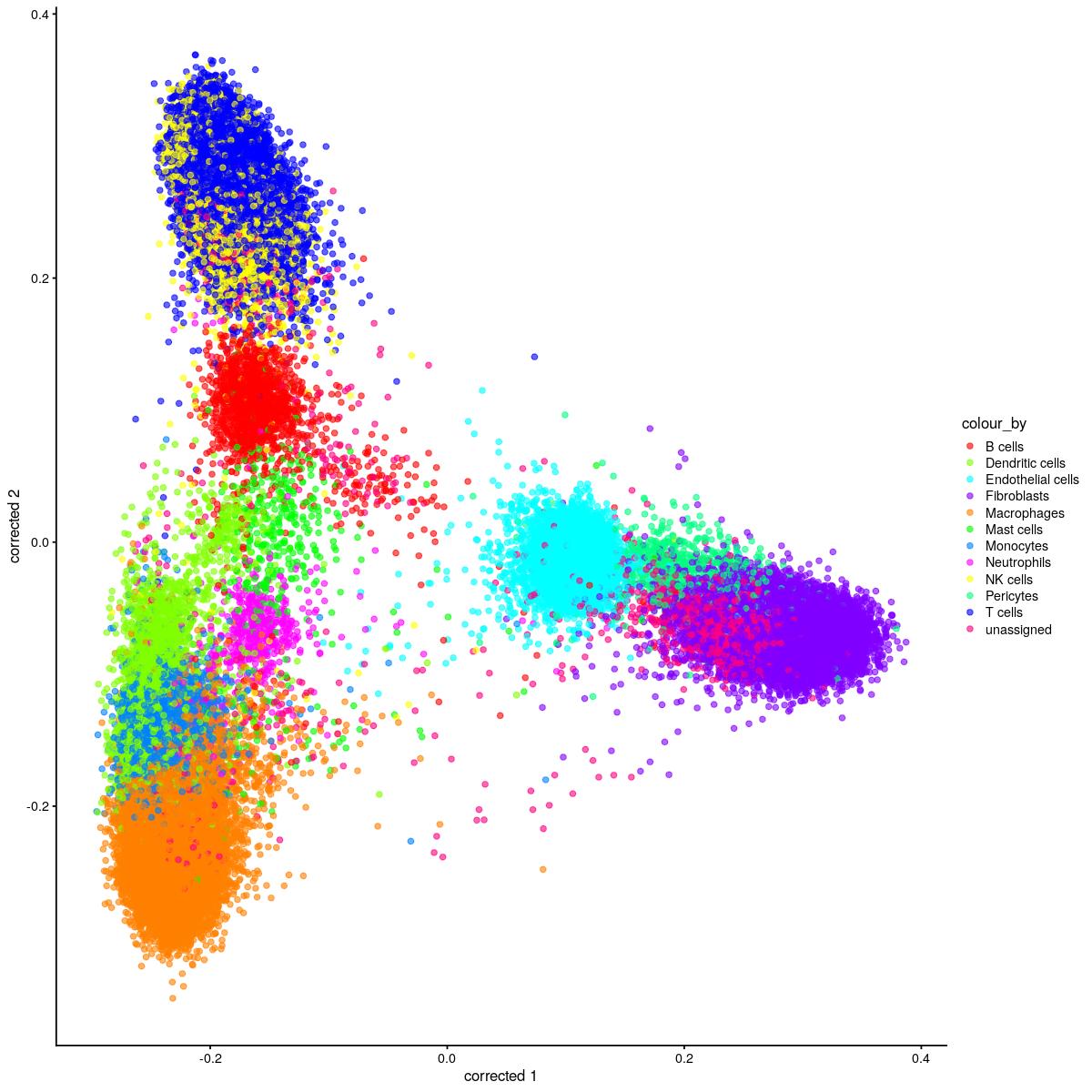
plotReducedDim(syn_sce_tidy_hvg_cms, "corrected", colour_by = "main_celltype_cellid") +
scale_color_manual(values = colors_used)Scale for 'colour' is already present. Adding another scale for 'colour',
which will replace the existing scale.
Past versions of plots_celltype_cellid-1.jpeg
Version
Author
Date
9133ed1
Reto Gerber
2022-03-04
222b0d1
Reto Gerber
2021-07-29
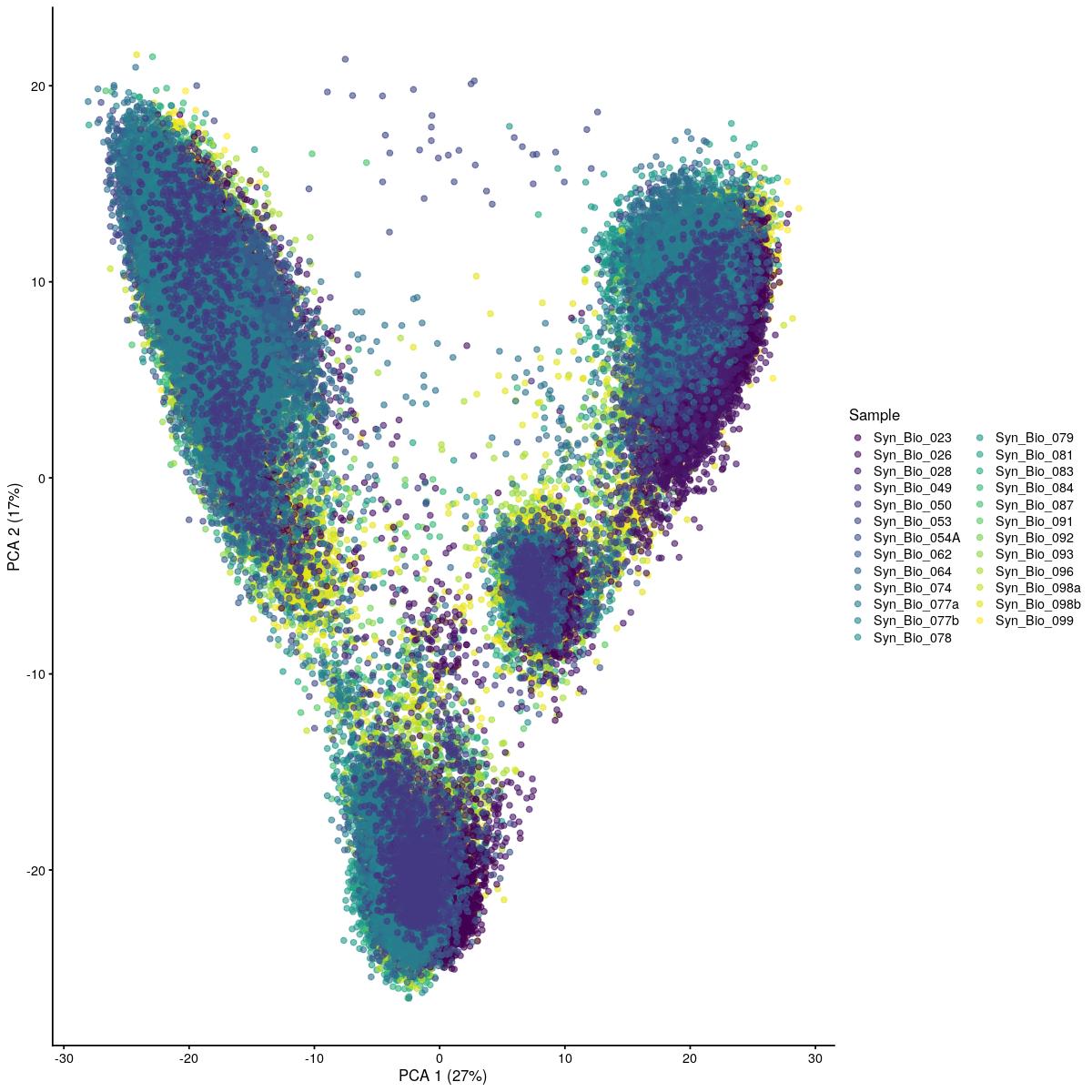
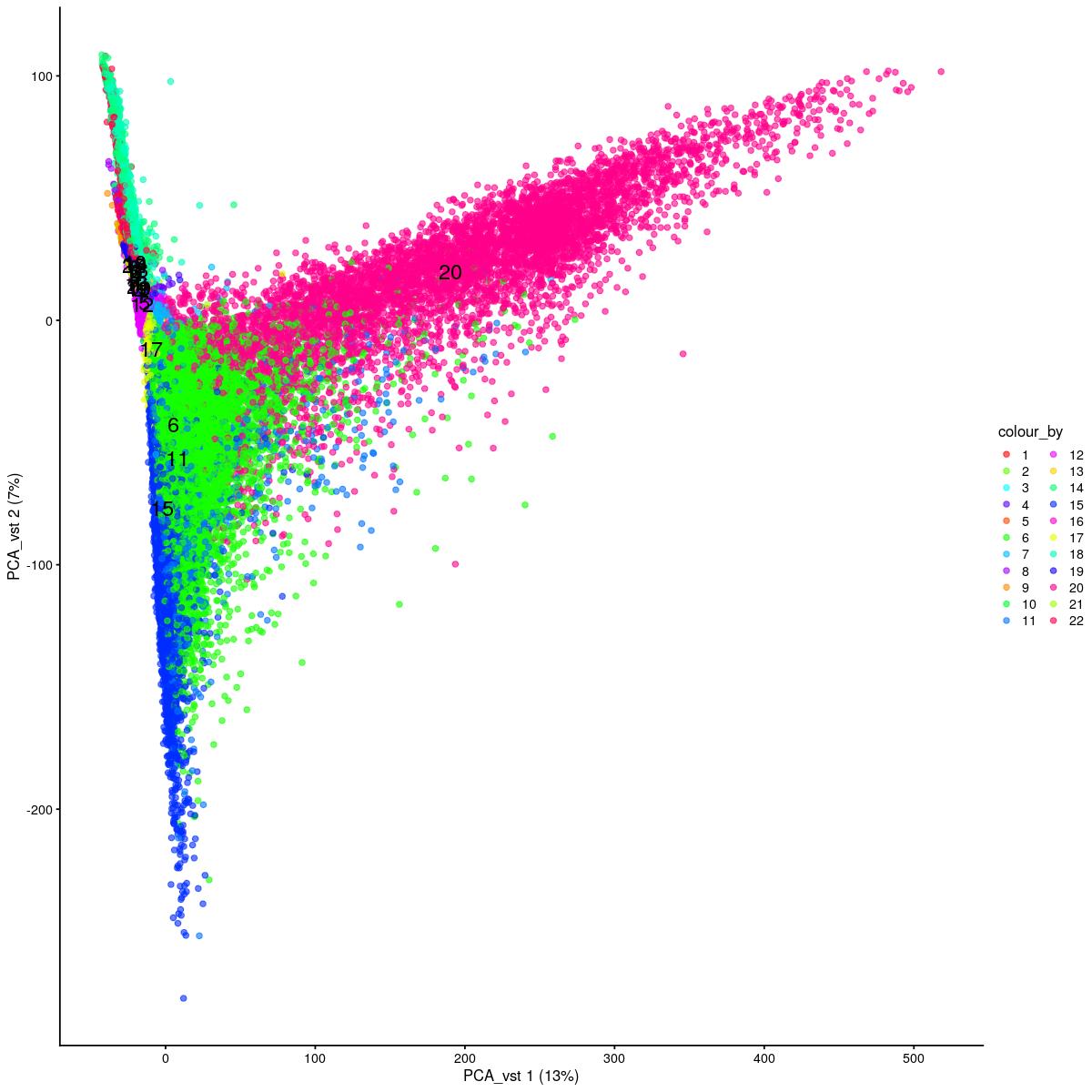
if(use_vst){
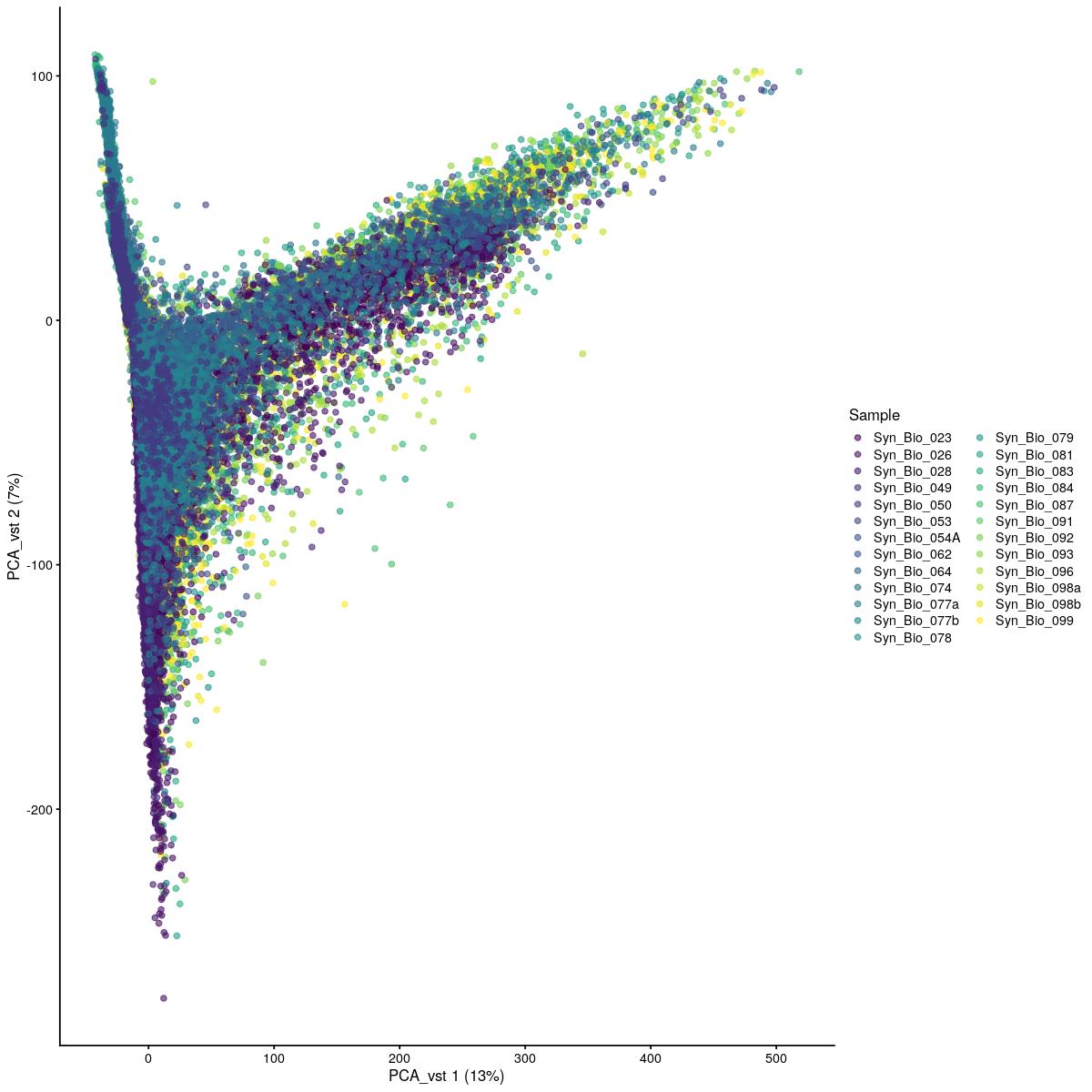
cat("\n\n#### uncorrected PCA vst\n\n")
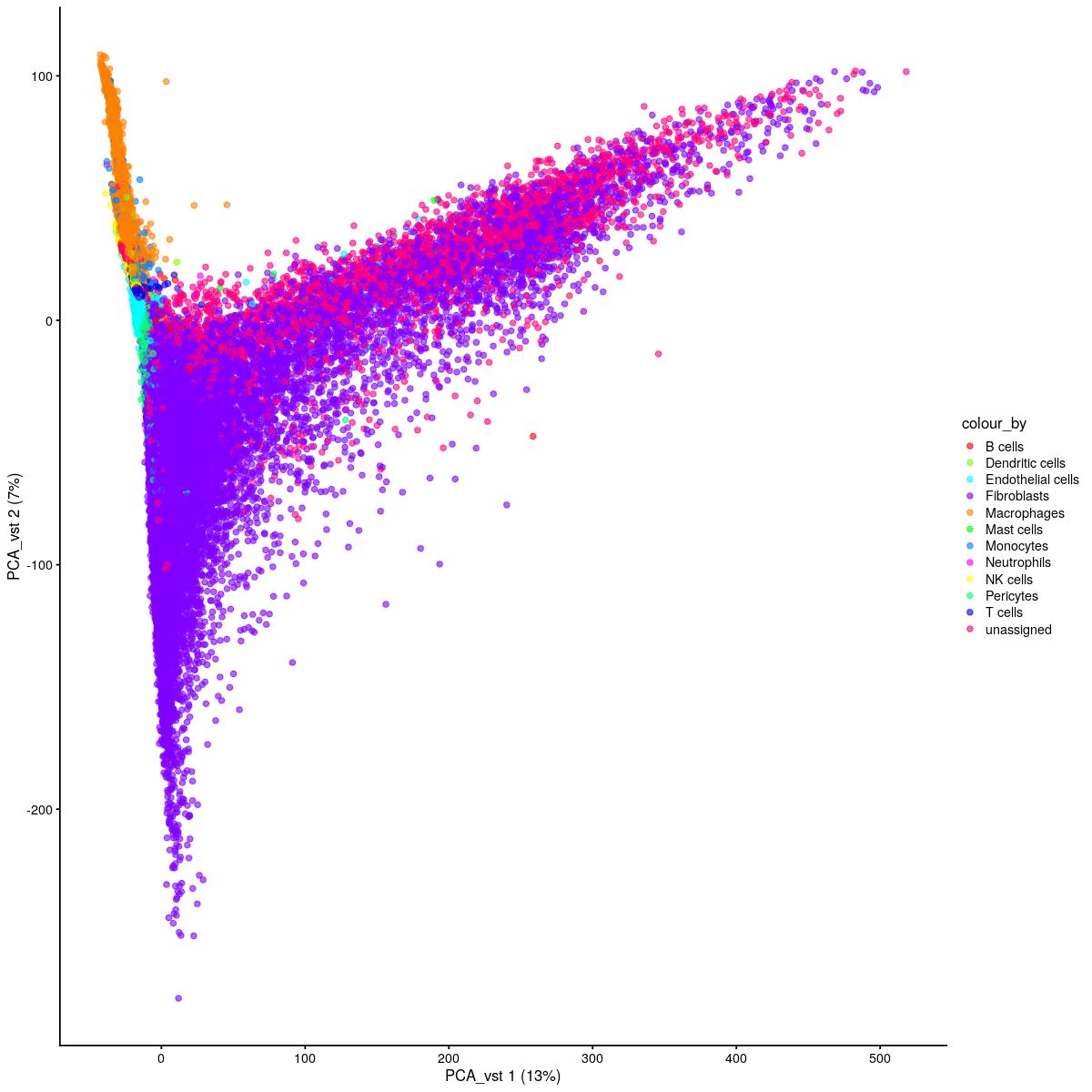
plotReducedDim(syn_sce_tidy_hvg_cms, "PCA_vst", colour_by = "main_celltype_cellid") +
scale_color_manual(values = colors_used)
}
uncorrected PCA vst
Scale for 'colour' is already present. Adding another scale for 'colour',
which will replace the existing scale.
Past versions of plots_celltype_cellid-2.jpeg
Version
Author
Date
9133ed1
Reto Gerber
2022-03-04
222b0d1
Reto Gerber
2021-07-29
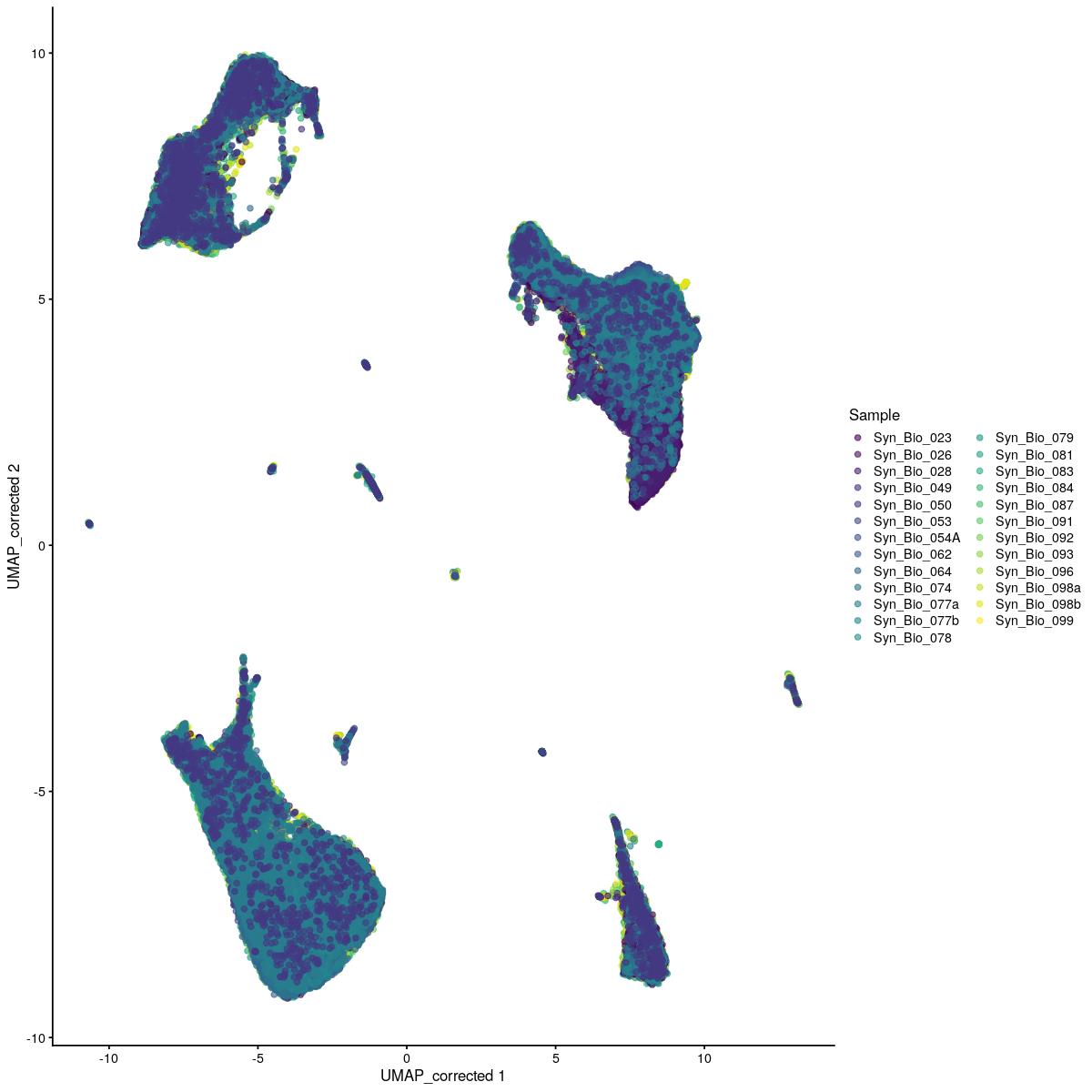
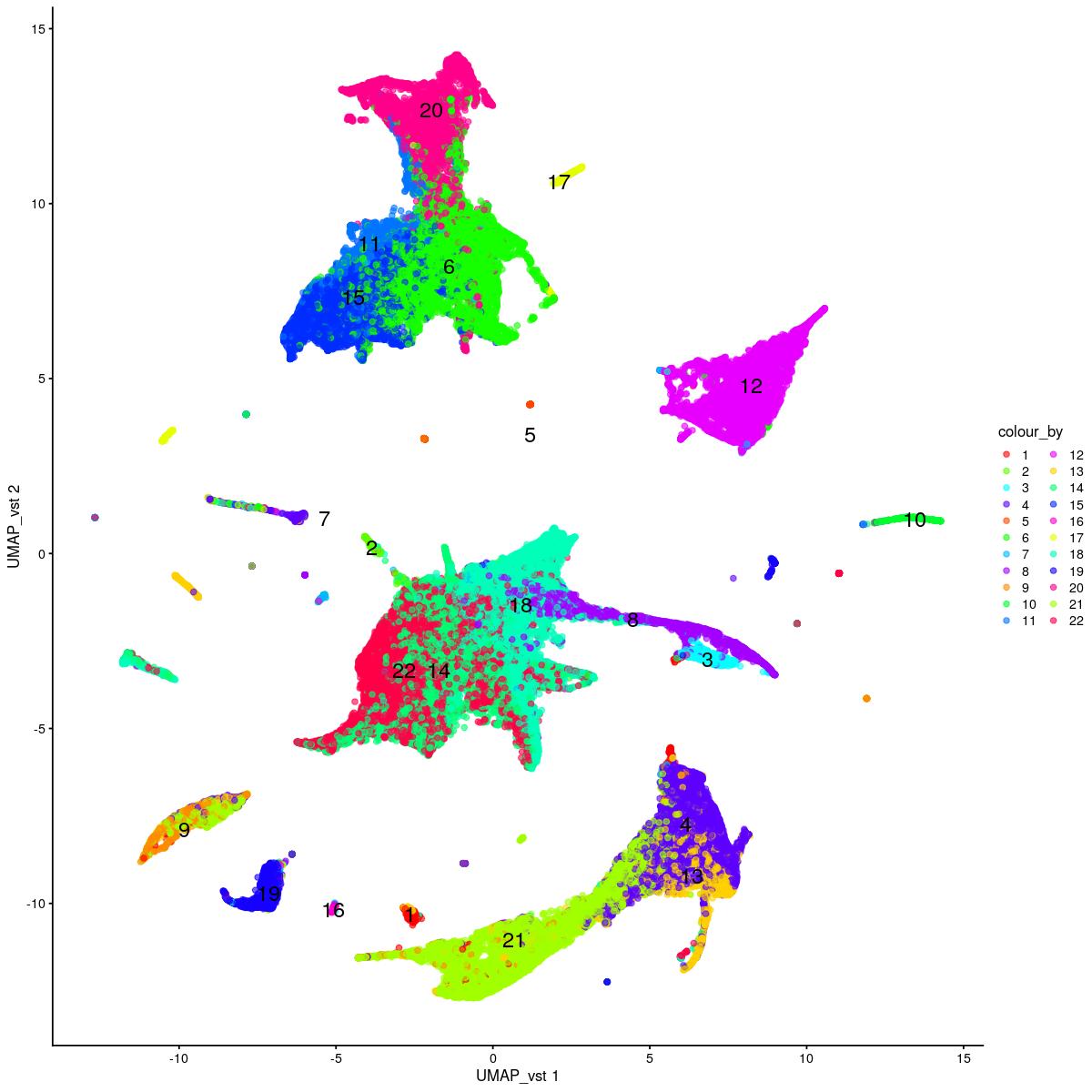
cat("\n\n#### corrected UMAP\n\n")
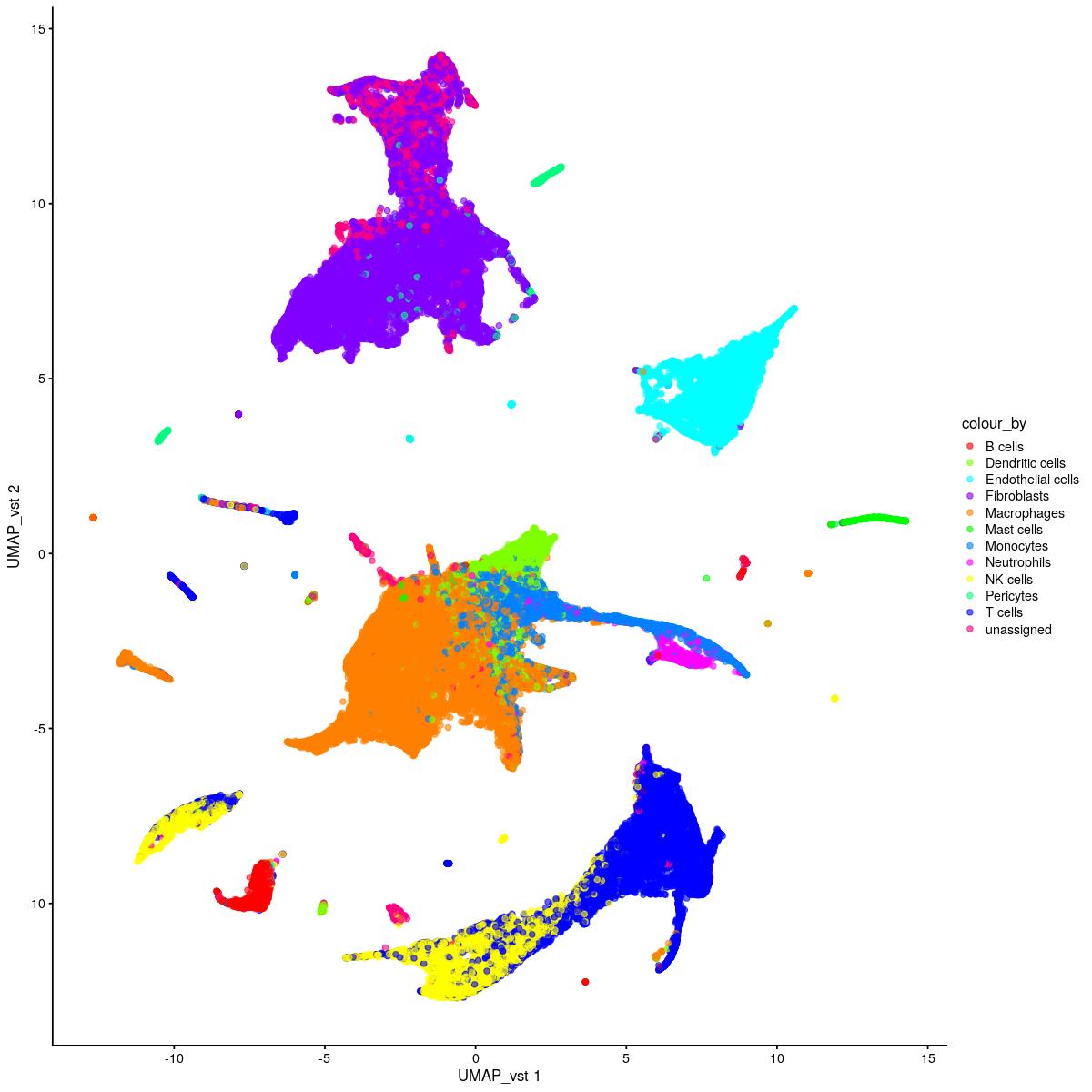
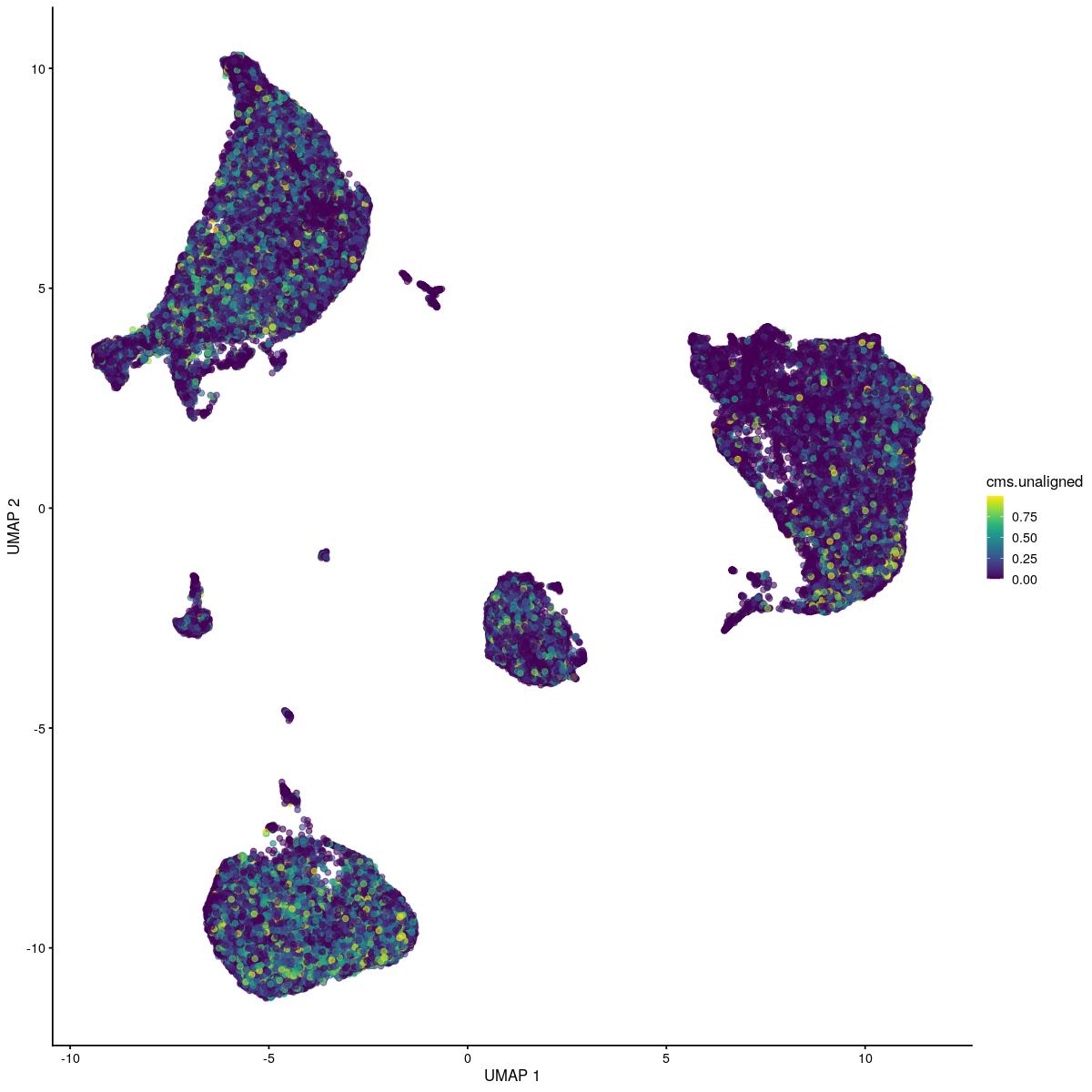
corrected UMAP
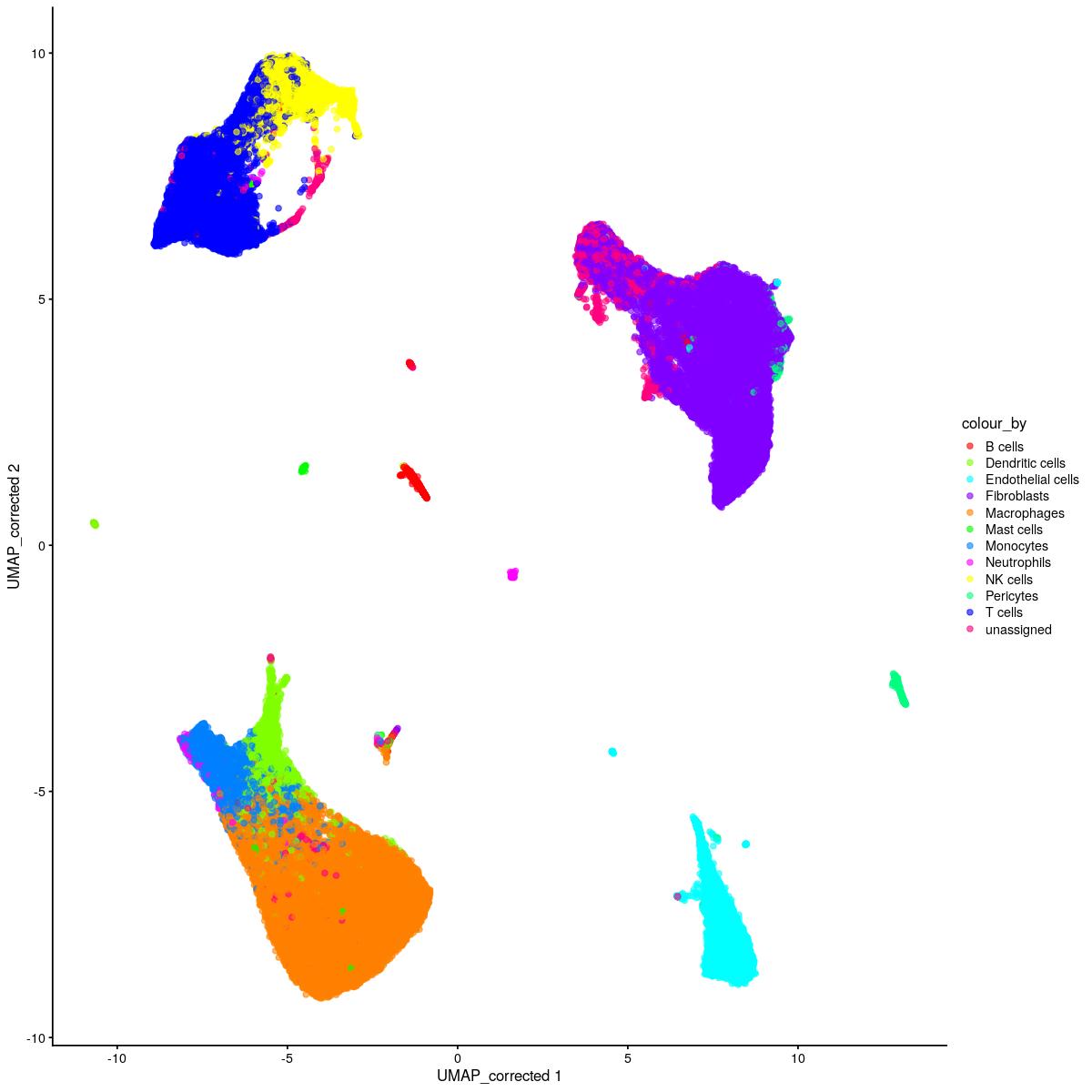
plotReducedDim(syn_sce_tidy_hvg_cms, "UMAP_corrected", colour_by = "main_celltype_cellid") +
scale_color_manual(values = colors_used)Scale for 'colour' is already present. Adding another scale for 'colour',
which will replace the existing scale.
Past versions of plots_celltype_cellid-3.jpeg
Version
Author
Date
9133ed1
Reto Gerber
2022-03-04
222b0d1
Reto Gerber
2021-07-29
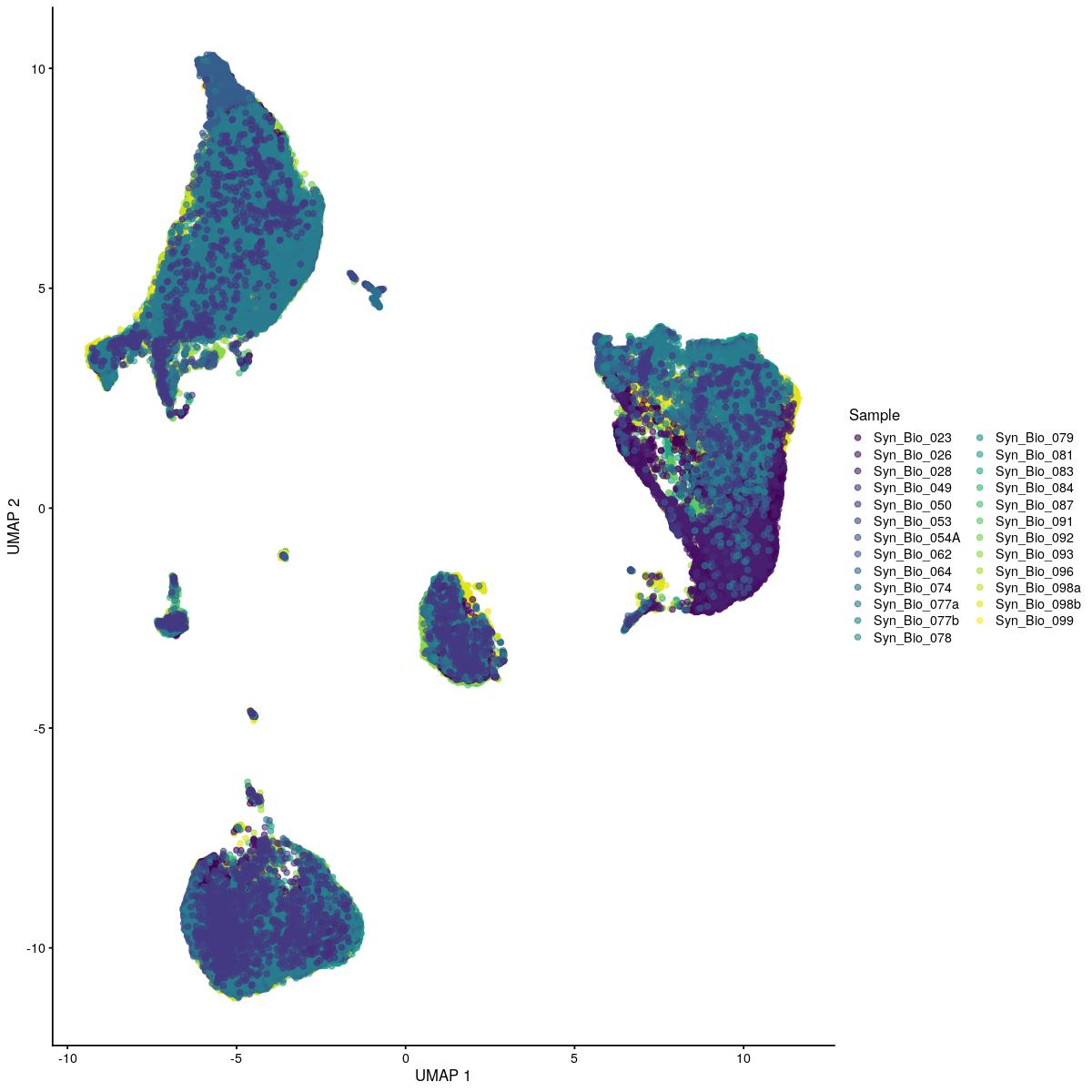
if(use_vst){
cat("\n\n#### uncorrected UMAP vst\n\n")
plotReducedDim(syn_sce_tidy_hvg_cms,"UMAP_vst", colour_by = "main_celltype_cellid") +
scale_color_manual(values = colors_used)
}
uncorrected UMAP vst
Scale for 'colour' is already present. Adding another scale for 'colour',
which will replace the existing scale.
Past versions of plots_celltype_cellid-4.jpeg
Version
Author
Date
9133ed1
Reto Gerber
2022-03-04
222b0d1
Reto Gerber
2021-07-29
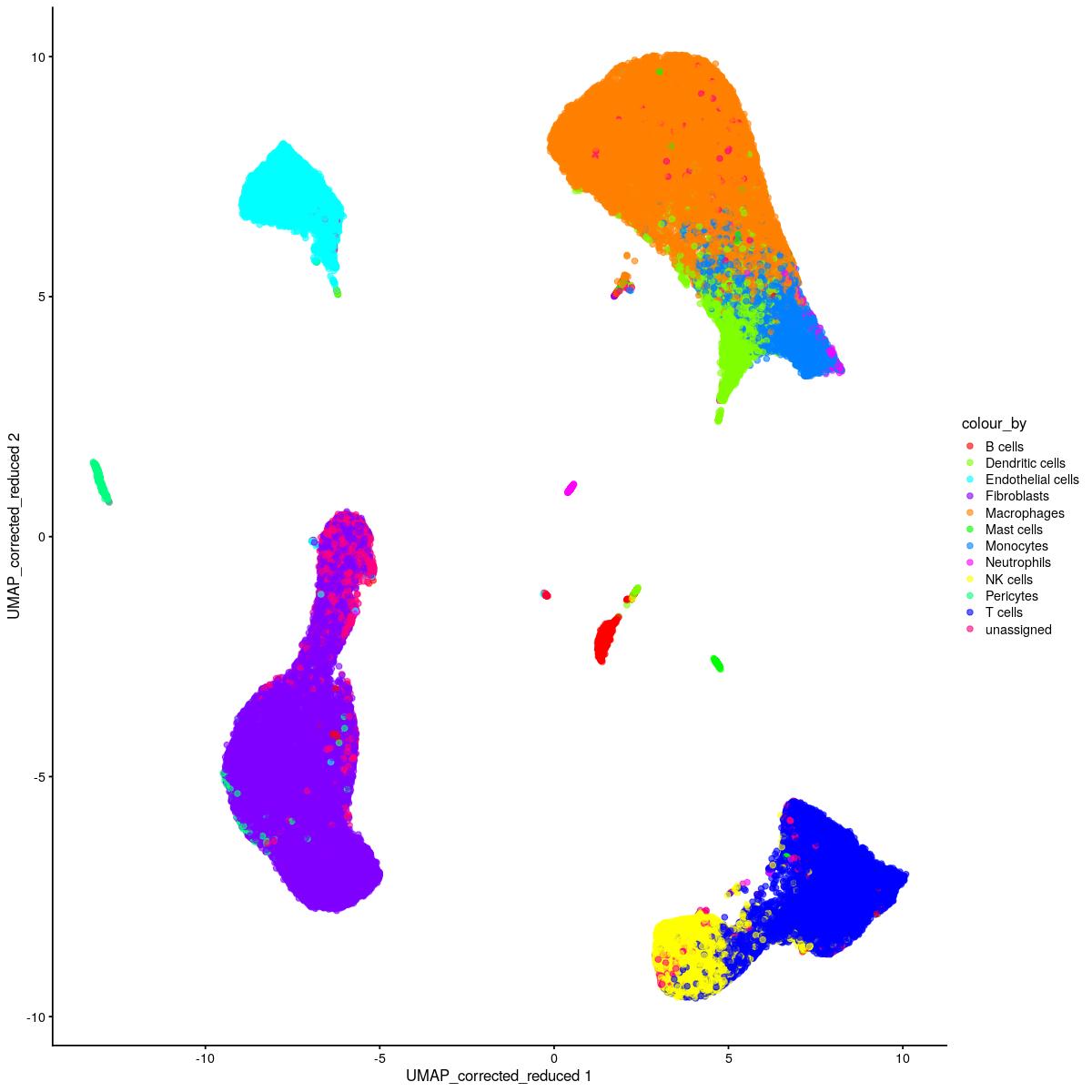
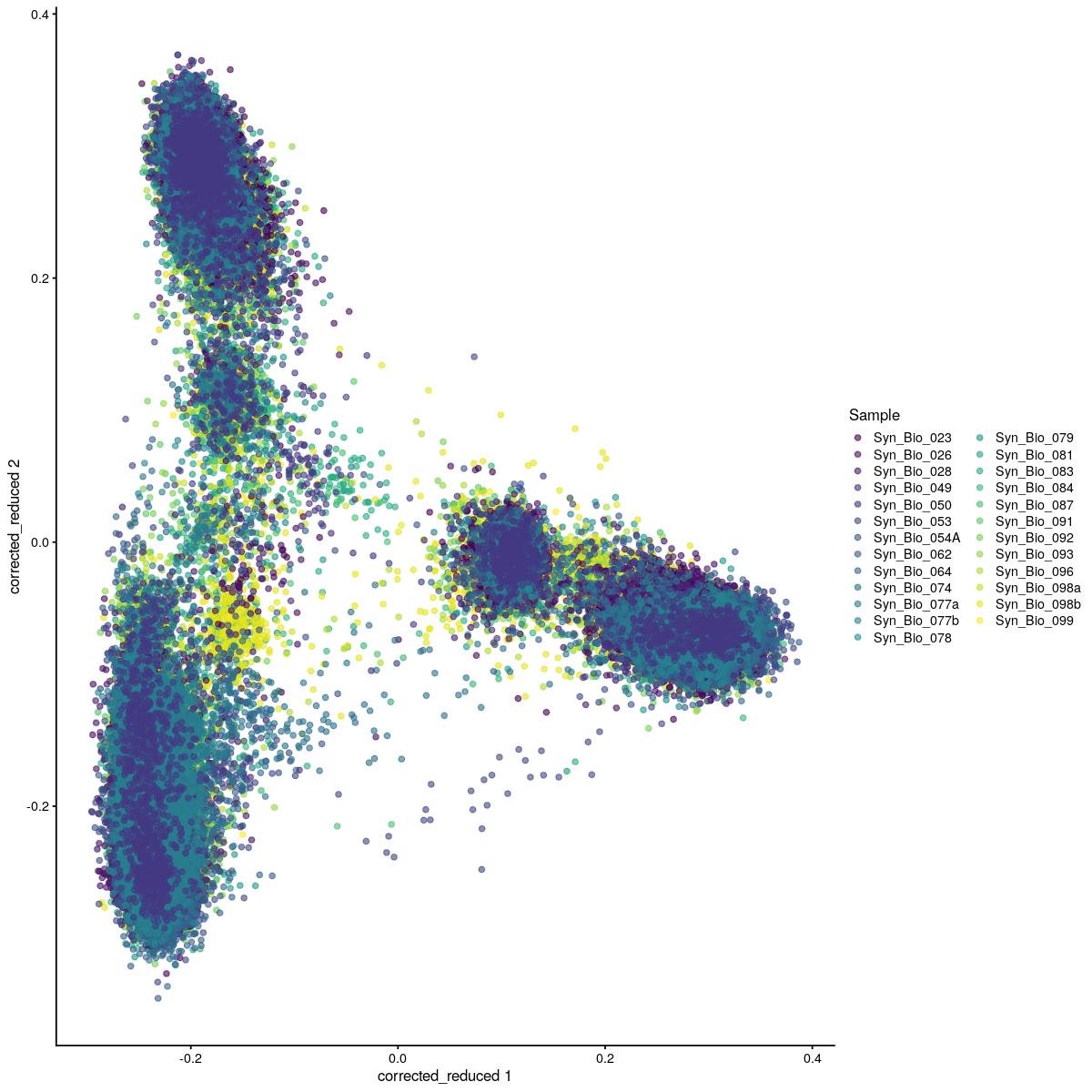
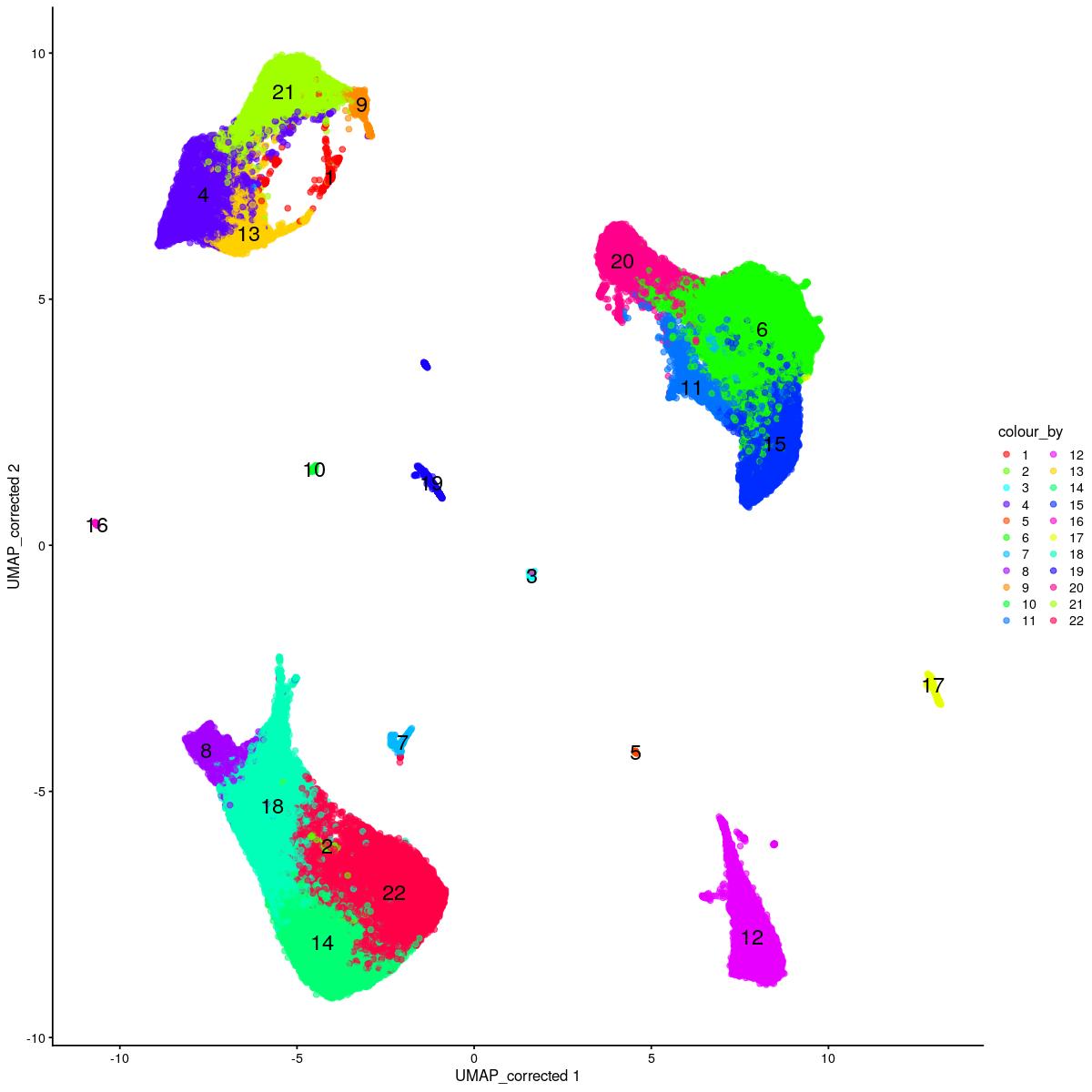
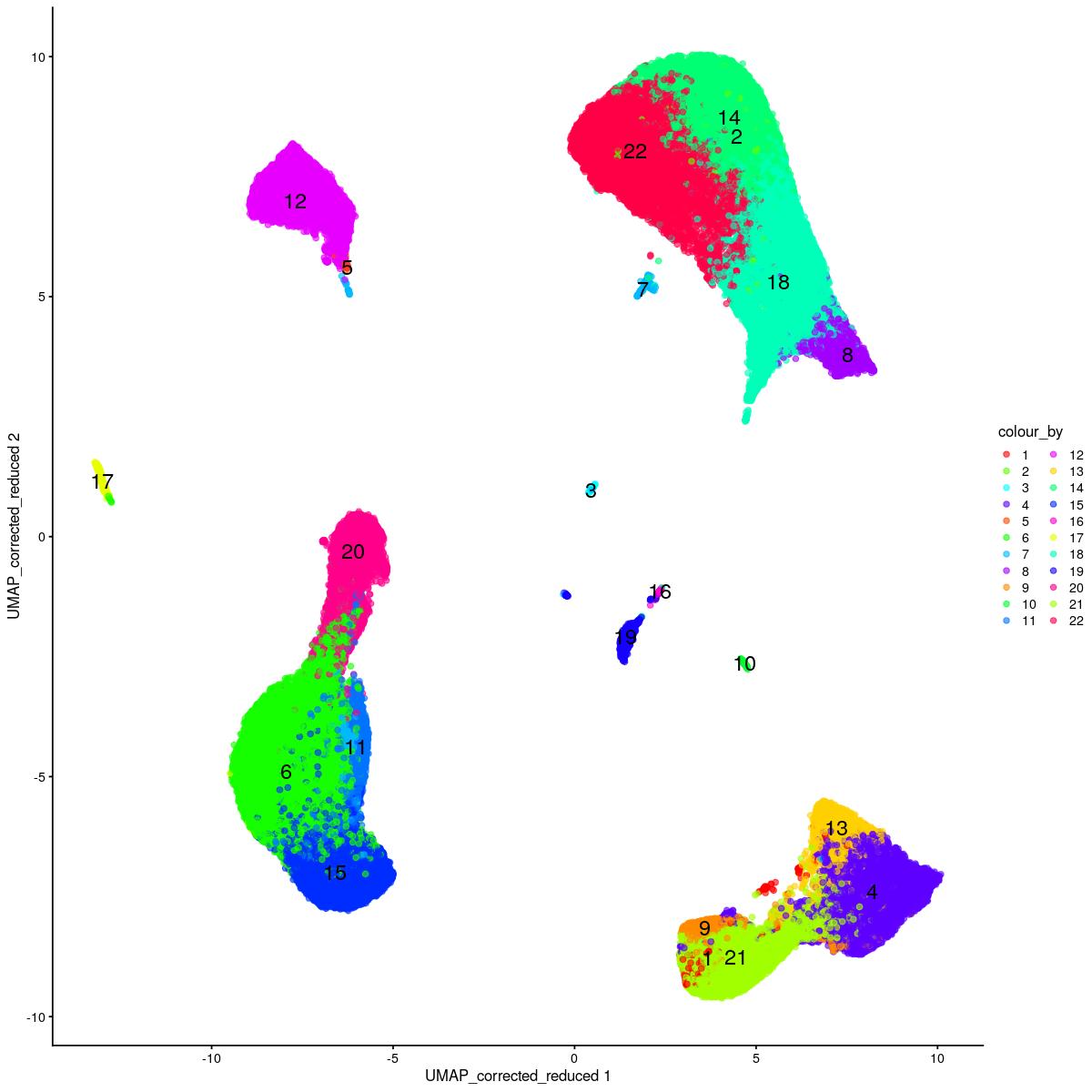
cat("\n\n#### corrected dimred UMAP\n\n")
corrected dimred UMAP
plotReducedDim(syn_sce_tidy_hvg_cms, "UMAP_corrected_reduced", colour_by = "main_celltype_cellid") +
scale_color_manual(values = colors_used)Scale for 'colour' is already present. Adding another scale for 'colour',
which will replace the existing scale.
Past versions of plots_celltype_cellid-5.jpeg
Version
Author
Date
9133ed1
Reto Gerber
2022-03-04
222b0d1
Reto Gerber
2021-07-29
cat("\n\n### {-}")